# library(tidyverse)
library(ggplot2)
library(vegan)
library(phyloseq)
# library (ape)
library(RColorBrewer)
Loading required package: permute
Loading required package: lattice
This is vegan 2.5-4
otu <- as.matrix(read.table("ASVs_counts.tsv", header=T, row.names=1)) #tabla de OTUs sin singletons, formato tabular. eliminados con: http://qiime.org/scripts/filter_otus_from_otu_table.html
OTU = otu_table(otu, taxa_are_rows=T)
head(OTU)
taximat = as.matrix(read.table("ASVs_taxonomy.tsv", header=T, row.names=1)) #revisar los encabezados
taxi=tax_table(taximat)
metro = phyloseq(OTU, taxi)
sample_names(metro)
data= read.table("THEmetadata.txt", header=T, row.names=1, sep="\t")#los metadatos deben de estar en el mismo orden en el que estan en la tabla de OTUs: sample_data(camaron)
sampledata = sample_data(data.frame(id=data$id, sample_location=data$sample_location, sample_id=data$sample_id,line_number=data$line_number, sample_type=data$sample_type, date=data$date, time=data$time, temperature_C=data$temperature_C, Humidity=data$Humidity, length_underground=data$ length_underground, length_superficial=data$length_superficial, length_elevated=data$length_elevated, length_total=data$length_total, elevation=data$elevation, station=data$station_hubs, train_track=data$train_track, train_notes=data$train_notes, station_notes=data$station_notes, geographical_zone=data$geographical_zone, weather=data$weather, stations_number=data$stations_number, latitude=data$latitude, longitude=data$longitude, people_affluence=data$people_affluence, Observed=data$Observed, Chao1=data$Chao1, Shannon=data$Shannon, Simpson=data$Simpson, row.names=sample_names(metro)))
# tree <- read.newick("metro.tre")
# tree <- collapse.singles(tree)
metro = phyloseq (OTU, sampledata, taxi)
metro
| <th scope=col>AM01</th><th scope=col>AM02</th><th scope=col>AM03</th><th scope=col>AM04</th><th scope=col>AM05</th><th scope=col>AM06</th><th scope=col>AM07</th><th scope=col>AM08</th><th scope=col>AM09</th><th scope=col>AM10</th><th scope=col>⋯</th><th scope=col>AM39</th><th scope=col>AM40</th><th scope=col>AM41</th><th scope=col>AM42</th><th scope=col>AM43</th><th scope=col>AM44</th><th scope=col>AM45</th><th scope=col>AM46</th><th scope=col>AM47</th><th scope=col>AM48</th> | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10459 | 8198 | 6740 | 5339 | 114 | 15981 | 9607 | 4123 | 5860 | 4780 | ⋯ | 9224 | 20148 | 15588 | 15573 | 7217 | 43753 | 19733 | 13121 | 35807 | 19170 |
| 2232 | 6735 | 3194 | 2244 | 69 | 3268 | 3663 | 1786 | 2574 | 3542 | ⋯ | 1476 | 3016 | 3018 | 1699 | 764 | 1450 | 3853 | 2261 | 1191 | 2595 |
| 997 | 1372 | 3194 | 0 | 0 | 0 | 0 | 462 | 0 | 0 | ⋯ | 487 | 389 | 299 | 3555 | 1510 | 477 | 0 | 3658 | 502 | 0 |
| 705 | 1456 | 454 | 1279 | 115 | 485 | 5619 | 367 | 2370 | 786 | ⋯ | 1682 | 3666 | 1036 | 2506 | 474 | 6183 | 5043 | 737 | 2413 | 2674 |
| 1752 | 1938 | 1800 | 1050 | 31 | 7149 | 1617 | 1268 | 1337 | 2806 | ⋯ | 2826 | 639 | 1106 | 1322 | 685 | 1740 | 2532 | 2583 | 690 | 4561 |
| 1053 | 2408 | 672 | 3084 | 72 | 1809 | 3995 | 675 | 7260 | 1097 | ⋯ | 3187 | 1961 | 1231 | 791 | 430 | 983 | 5016 | 1659 | 305 | 1469 |
<ol class=list-inline> <li>‘AM01’</li> <li>‘AM02’</li> <li>‘AM03’</li> <li>‘AM04’</li> <li>‘AM05’</li> <li>‘AM06’</li> <li>‘AM07’</li> <li>‘AM08’</li> <li>‘AM09’</li> <li>‘AM10’</li> <li>‘AM11’</li> <li>‘AM12’</li> <li>‘AM13’</li> <li>‘AM14’</li> <li>‘AM15’</li> <li>‘AM16’</li> <li>‘AM17’</li> <li>‘AM18’</li> <li>‘AM19’</li> <li>‘AM20’</li> <li>‘AM21’</li> <li>‘AM22’</li> <li>‘AM23’</li> <li>‘AM24’</li> <li>‘AM26’</li> <li>‘AM27’</li> <li>‘AM28’</li> <li>‘AM29’</li> <li>‘AM30’</li> <li>‘AM31’</li> <li>‘AM32’</li> <li>‘AM33’</li> <li>‘AM34’</li> <li>‘AM35’</li> <li>‘AM36’</li> <li>‘AM37’</li> <li>‘AM38’</li> <li>‘AM39’</li> <li>‘AM40’</li> <li>‘AM41’</li> <li>‘AM42’</li> <li>‘AM43’</li> <li>‘AM44’</li> <li>‘AM45’</li> <li>‘AM46’</li> <li>‘AM47’</li> <li>‘AM48’</li> </ol>
phyloseq-class experiment-level object
otu_table() OTU Table: [ 22673 taxa and 47 samples ]
sample_data() Sample Data: [ 47 samples by 28 sample variables ]
tax_table() Taxonomy Table: [ 22673 taxa by 6 taxonomic ranks ]
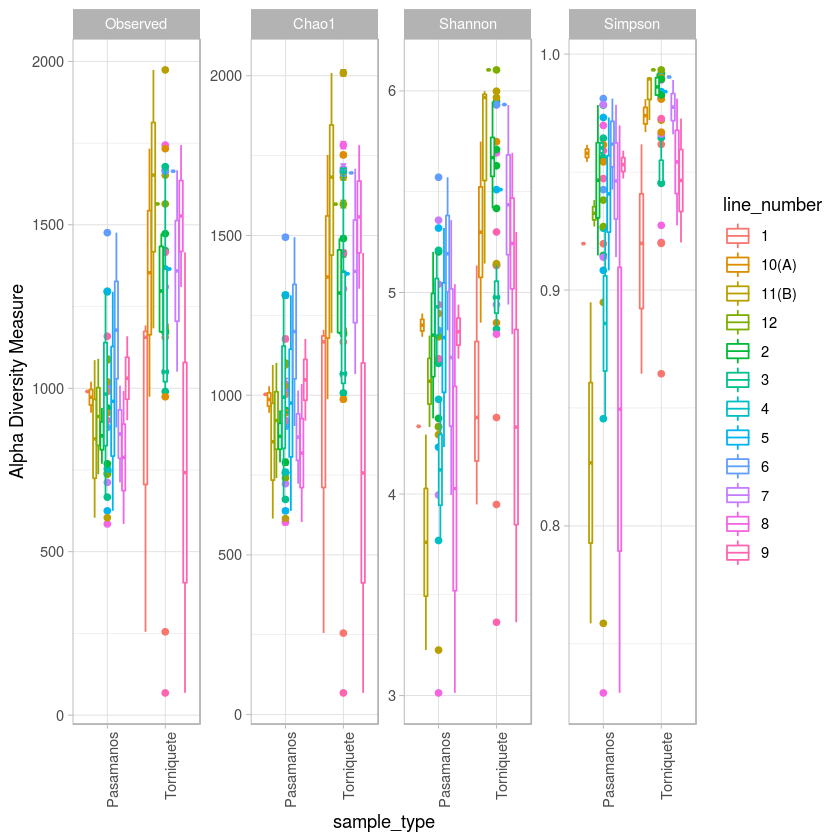
p <- plot_richness(metro, x="sample_type", color="line_number", measures=c("Observed", "Chao1", "Shannon", "Simpson")) + geom_boxplot()+theme_light() + theme(axis.text.x=element_text(angle=90, hjust=1))
#p$data
a <-p$data
p
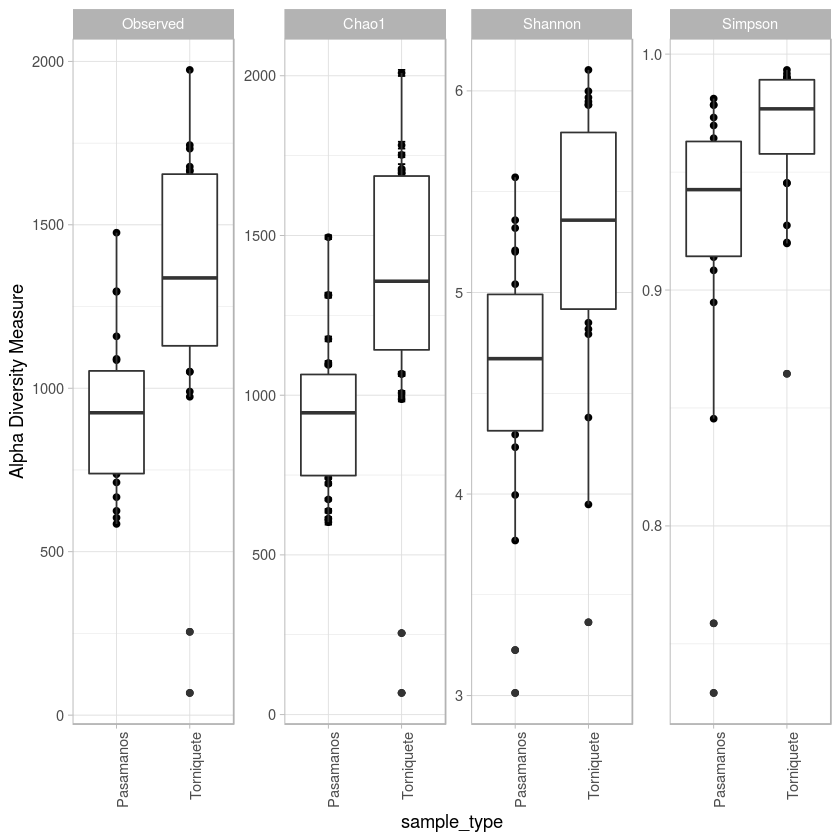
p <- plot_richness(metro, x="sample_type", measures=c("Observed", "Chao1", "Shannon", "Simpson")) + geom_boxplot()+theme_light() + theme(axis.text.x=element_text(angle=90, hjust=1))
#p$data
p
ggsave("histogramaASV_Alfa_diversidad.pdf")
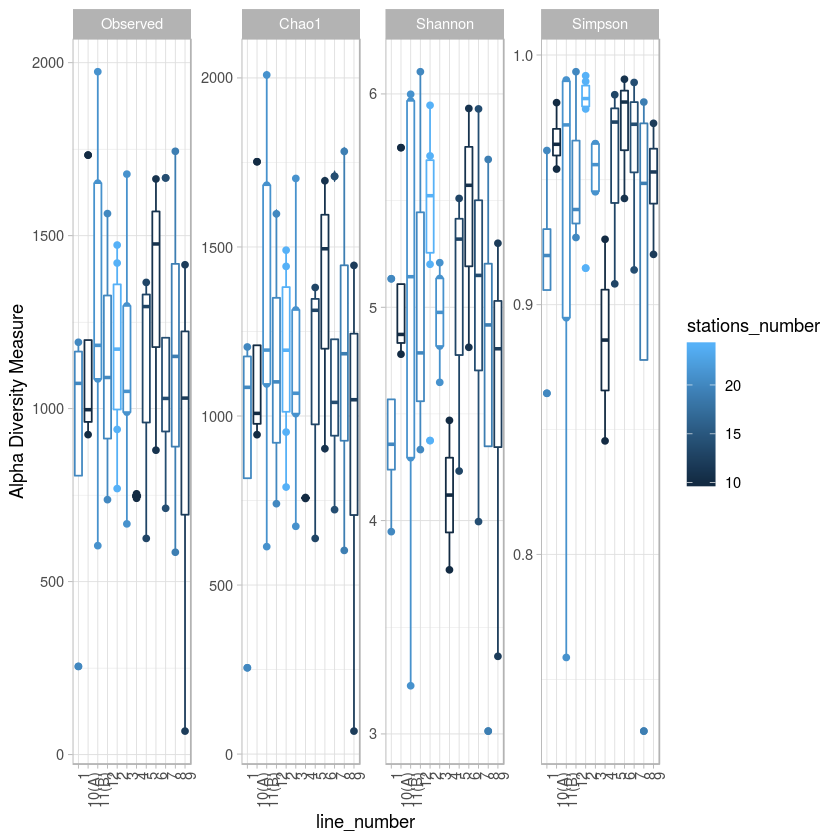
p <- plot_richness(metro, x="line_number", color="stations_number", measures=c("Observed", "Chao1", "Shannon", "Simpson")) + geom_boxplot()+theme_light() + theme(axis.text.x=element_text(angle=90, hjust=1))
#p$data
p
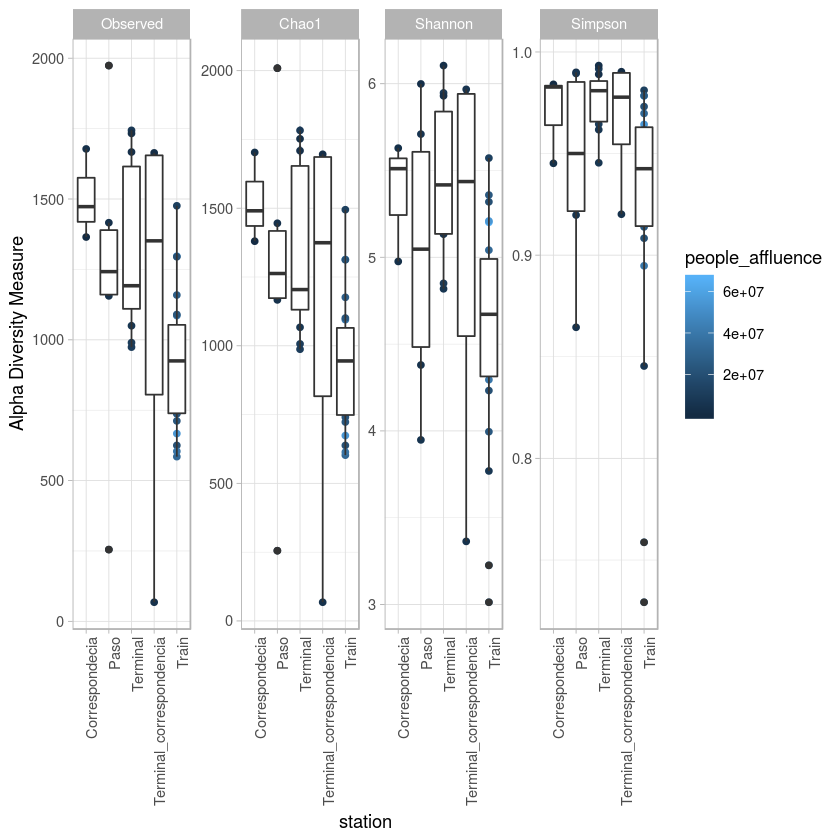
p <- plot_richness(metro, x="station",color="people_affluence", measures=c("Observed", "Chao1", "Shannon", "Simpson")) + geom_boxplot()+theme_light() + theme(axis.text.x=element_text(angle=90, hjust=1))
#p$data
p
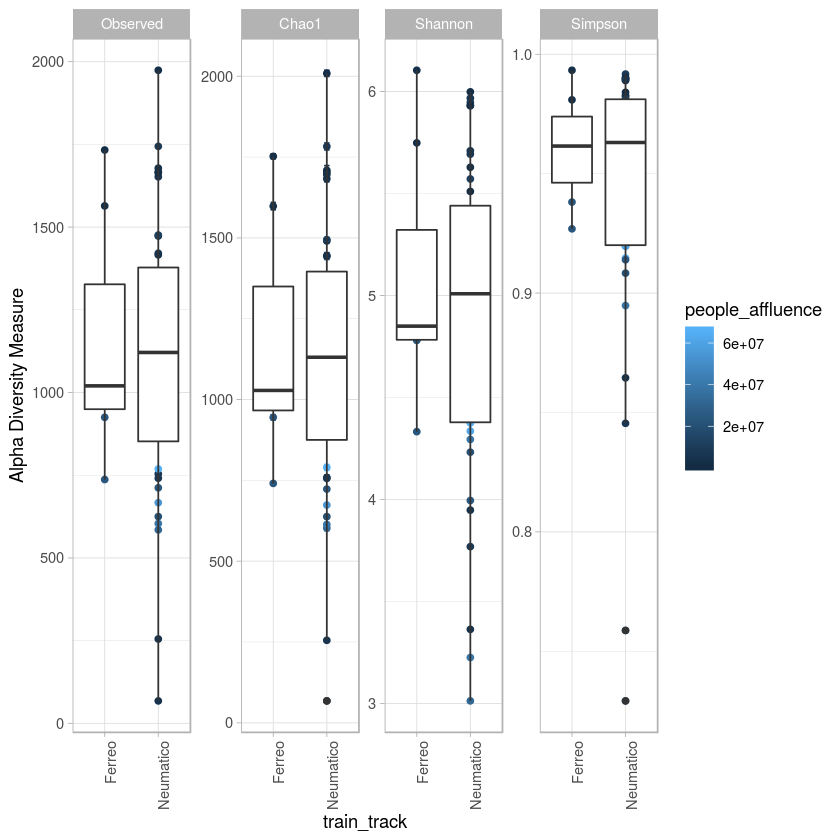
p <- plot_richness(metro, x="train_track",color="people_affluence", measures=c("Observed", "Chao1", "Shannon", "Simpson")) + geom_boxplot()+theme_light() + theme(axis.text.x=element_text(angle=90, hjust=1))
#p$data
p
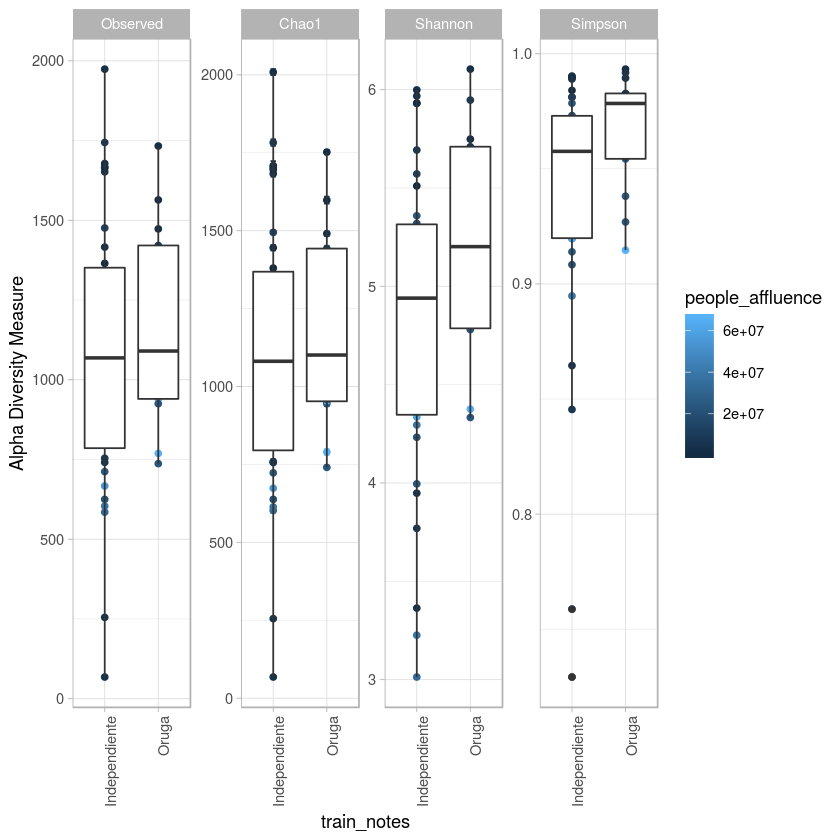
p <- plot_richness(metro, x="train_notes",color="people_affluence", measures=c("Observed", "Chao1", "Shannon", "Simpson")) + geom_boxplot()+theme_light() + theme(axis.text.x=element_text(angle=90, hjust=1))
#p$data
p
p <- plot_richness(metro, x="station_notes",color="people_affluence", measures=c("Observed", "Chao1", "Shannon", "Simpson")) + geom_boxplot()+theme_light() + theme(axis.text.x=element_text(angle=90, hjust=1))
#p$data
# p
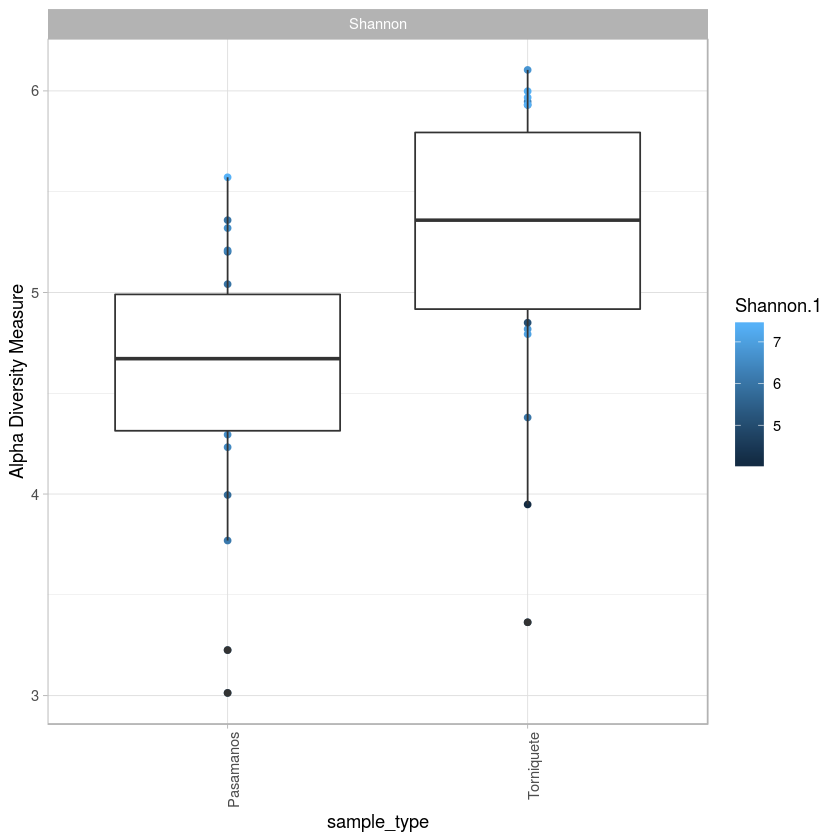
p <- plot_richness(metro, x="sample_type",color="Shannon.1", measures= "Shannon") + geom_boxplot()+theme_light() + theme(axis.text.x=element_text(angle=90, hjust=1))
a<- p$data
summary
p
write.csv(a,"shannon.estacion.csv",sep="\t")
id sample_location sample_id
Barranca_del_muerto_7 : 4 Train :92 AM01 : 4
Buenavista_B : 4 Barranca_del_muerto : 4 AM02 : 4
Chilpancingo_9 : 4 Buenavista : 4 AM03 : 4
Ciudad_Azteca_B : 4 Chilpancingo : 4 AM04 : 4
Constitucion_de_1917_8: 4 Ciudad_Azteca : 4 AM05 : 4
Cuatro_caminos_2 : 4 Constitucion_de_1917: 4 AM06 : 4
(Other) :164 (Other) :76 (Other):164
line_number sample_type date time temperature_C
2 :24 Pasamanos :92 2/5/16 :36 11:25:00 AM: 8 Min. :25.90
11(B) :20 Torniquete:96 3/5/16 :28 11:42:00 AM: 8 1st Qu.:29.10
3 :20 4/16/27:36 1:20:00 PM : 8 Median :30.40
1 :16 4/16/28:24 12:05:00 PM: 8 Mean :30.15
10(A) :16 4/16/29:40 12:40:00 PM: 8 3rd Qu.:31.30
7 :16 4/5/16 :24 12:50:00 PM: 8 Max. :33.30
(Other):76 (Other) :140
Humidity length_underground length_superficial length_elevated
Min. :25.00 Min. : 0.00 Min. : 0.000 Min. : 0.000
1st Qu.:27.00 1st Qu.: 5.38 1st Qu.: 0.000 1st Qu.: 0.000
Median :30.00 Median :11.86 Median : 4.449 Median : 0.000
Mean :30.38 Mean :11.05 Mean : 5.485 Mean : 2.001
3rd Qu.:32.00 3rd Qu.:16.79 3rd Qu.:10.724 3rd Qu.: 4.185
Max. :41.00 Max. :18.14 Max. :15.151 Max. :11.533
length_total elevation station train_track
Min. :11.00 Min. :2231 Correspondecia :12 Ferreo : 28
1st Qu.:16.00 1st Qu.:2237 Paso :24 Neumatico:160
Median :18.00 Median :2242 Terminal :44
Mean :18.83 Mean :2250 Terminal_correspondencia:16
3rd Qu.:22.00 3rd Qu.:2256 Train :92
Max. :25.00 Max. :2303
NA's :92
train_notes station_notes geographical_zone weather
Independiente:136 Cajon_subterraneo:44 Centro :20 B1 :16
Oruga : 52 Elevada :36 Norte :28 C1 :64
Superficial :12 Oriente :20 C2 :16
Tunel : 4 Poniente:12 NA's:92
NA's :92 Sur :16
NA's :92
stations_number latitude longitude people_affluence
Min. :10.00 Min. :19.29 Min. :-99.22 Min. : 499350
1st Qu.:12.00 1st Qu.:19.39 1st Qu.:-99.17 1st Qu.: 4190568
Median :19.00 Median :19.43 Median :-99.14 Median : 9523016
Mean :17.15 Mean :19.43 Mean :-99.13 Mean :17859153
3rd Qu.:21.00 3rd Qu.:19.49 3rd Qu.:-99.10 3rd Qu.:24196452
Max. :24.00 Max. :19.53 Max. :-98.96 Max. :66232325
NA's :92 NA's :92
Observed.1 Chao1.1 Shannon.1 Simpson.1
Min. : 428 Min. : 840.2 Min. :4.102 Min. :0.8984
1st Qu.: 7655 1st Qu.:12618.1 1st Qu.:6.004 1st Qu.:0.9802
Median :10129 Median :15375.8 Median :6.410 Median :0.9872
Mean : 9904 Mean :15141.1 Mean :6.380 Mean :0.9827
3rd Qu.:12677 3rd Qu.:18521.1 3rd Qu.:6.863 3rd Qu.:0.9916
Max. :16179 Max. :23052.1 Max. :7.384 Max. :0.9960
samples variable value se
Length:188 Observed:47 Min. : 0.7292 Min. : 0.000
Class :character Chao1 :47 1st Qu.: 2.5084 1st Qu.: 5.580
Mode :character Shannon :47 Median : 37.0522 Median : 7.086
Simpson :47 Mean : 561.0525 Mean : 6.989
3rd Qu.:1087.0000 3rd Qu.: 8.543
Max. :2009.0065 Max. :17.183
NA's :141
Warning message:
“Removed 141 rows containing missing values (geom_errorbar).”Warning message:
“Removed 141 rows containing missing values (geom_errorbar).”
Saving 6.67 x 6.67 in image
Warning message:
“Removed 141 rows containing missing values (geom_errorbar).”Warning message:
“Removed 141 rows containing missing values (geom_errorbar).”
Warning message:
“Removed 141 rows containing missing values (geom_errorbar).”
Warning message:
“Removed 141 rows containing missing values (geom_errorbar).”
Warning message:
“Removed 141 rows containing missing values (geom_errorbar).”
Warning message in write.csv(a, "shannon.estacion.csv", sep = "\t"):
“attempt to set 'sep' ignored”
library(ggrepel)
p <- plot_richness(metro, x="sample_type",color="Shannon.1", measures= "Shannon")
p + geom_point(size=5, alpha=1) + scale_fill_gradient(low="#F1E8FA", high="#FA7500", na.value = "white") + theme_light() + theme(axis.text.x=element_text(angle=90, hjust=1))
a<-as.data.frame(p$data)
head(a)
# head(p$data$id)
# head(a)
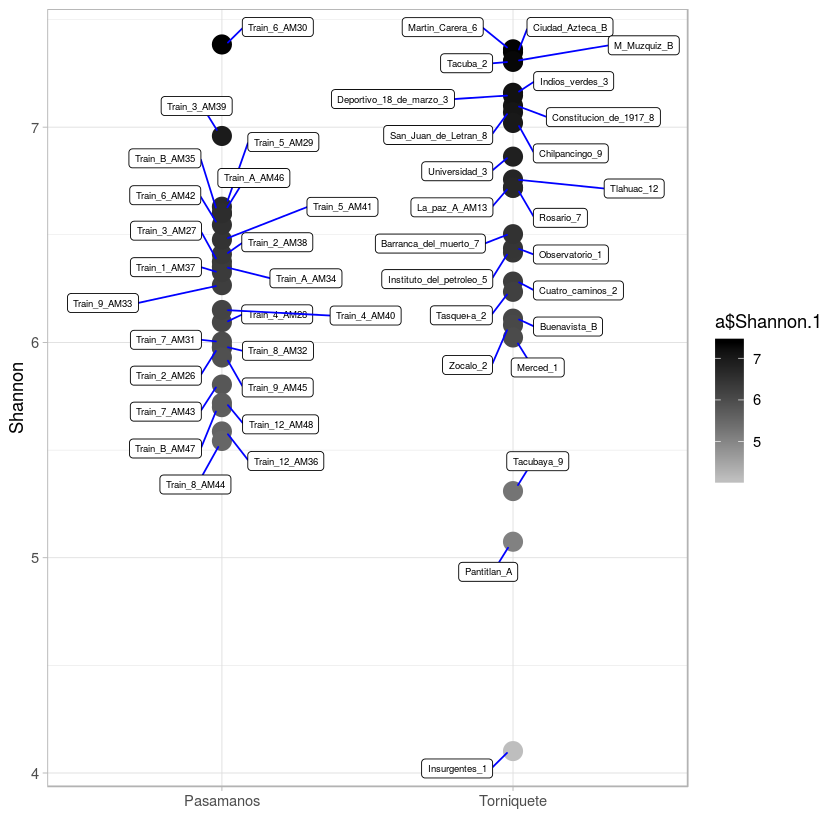
shannonplot <- ggplot(a, aes(x=a$sample_type, y=(a$Shannon.1))) + geom_point(aes(colour=a$Shannon.1), size=5) + scale_colour_gradient(low="#bebebe", high="#000000", na.value = "white")+ theme_light() +
# geom_text(aes(label=a$id), hjust=-0.1, vjust=0, size=2) +
xlab("") + ylab("Shannon")
shannonplot + geom_label_repel(aes(label = a$id),
box.padding = 0.35,
point.padding = 0.5,
segment.color = 'blue', size=2)
# ggplot(data=yx, aes(x = yx$orden, y= reorder(Genus, Abundance), fill= Abundance)) +
# geom_raster() + theme_minimal () +
# xlab("") + ylab("") +
# scale_fill_gradient(low="#bebebe", high="#000000", na.value = "white", trans = "log10") +
# theme(axis.text.x = element_text( angle = 90, hjust = 1),
# axis.text.y = element_text(size = 4))
# write.csv(a,"shannon.estacion.csv",sep="\t")
# library(ggplot2)
# library(ggrepel)
# nba <- read.csv("http://datasets.flowingdata.com/ppg2008.csv", sep = ",")
# nbaplot <- ggplot(nba, aes(x= MIN, y = PTS)) +
# geom_point(color = "blue", size = 3)
# ### geom_label_repel
# nbaplot +
# geom_label_repel(aes(label = Name),
# box.padding = 0.35,
# point.padding = 0.5,
# segment.color = 'grey50') +
# theme_classic()
| Indios_verdes_3 | Indios_verdes | AM01 | 3 | Torniquete | 2/5/16 | 2:23:00 PM | 30.4 | 27 | 18.145 | ⋯ | -99.11940 | 10176457 | 12677 | 19699.2193 | 7.160039 | 0.9921556 | AM01 | Shannon | 5.135283 | NA |
| Instituto_del_petroleo_5 | Instituto_del_petroleo | AM02 | 5 | Torniquete | 4/16/29 | 1:17:00 PM | 28.3 | 30 | 4.951 | ⋯ | -99.14480 | 499350 | 10722 | 16496.3811 | 6.417885 | 0.9890120 | AM02 | Shannon | 5.510396 | NA |
| Rosario_7 | Rosario | AM03 | 7 | Torniquete | 4/16/29 | 12:40:00 PM | 29.2 | 29 | 17.754 | ⋯ | -99.17970 | 3220719 | 11986 | 18521.1304 | 6.718174 | 0.9892510 | AM03 | Shannon | 4.939963 | NA |
| Cuatro_caminos_2 | Cuatro_caminos | AM04 | 2 | Torniquete | 4/16/29 | 11:42:00 AM | 30.3 | 30 | 12.550 | ⋯ | -99.21550 | 9523016 | 5425 | 8341.2612 | 6.283001 | 0.9939737 | AM04 | Shannon | 5.946868 | NA |
| Tacubaya_9 | Tacubaya | AM05 | 9 | Torniquete | 4/16/27 | 1:41:00 PM | 30.4 | 26 | 9.531 | ⋯ | -99.18830 | 4190568 | 428 | 840.2462 | 5.309640 | 0.9899266 | AM05 | Shannon | 3.363853 | NA |
| Observatorio_1 | Observatorio | AM06 | 1 | Torniquete | 4/5/16 | 2:35:00 PM | 32.2 | 28 | 16.786 | ⋯ | -99.19978 | 6489055 | 10523 | 17035.5115 | 6.437592 | 0.9859032 | AM06 | Shannon | 5.133815 | NA |
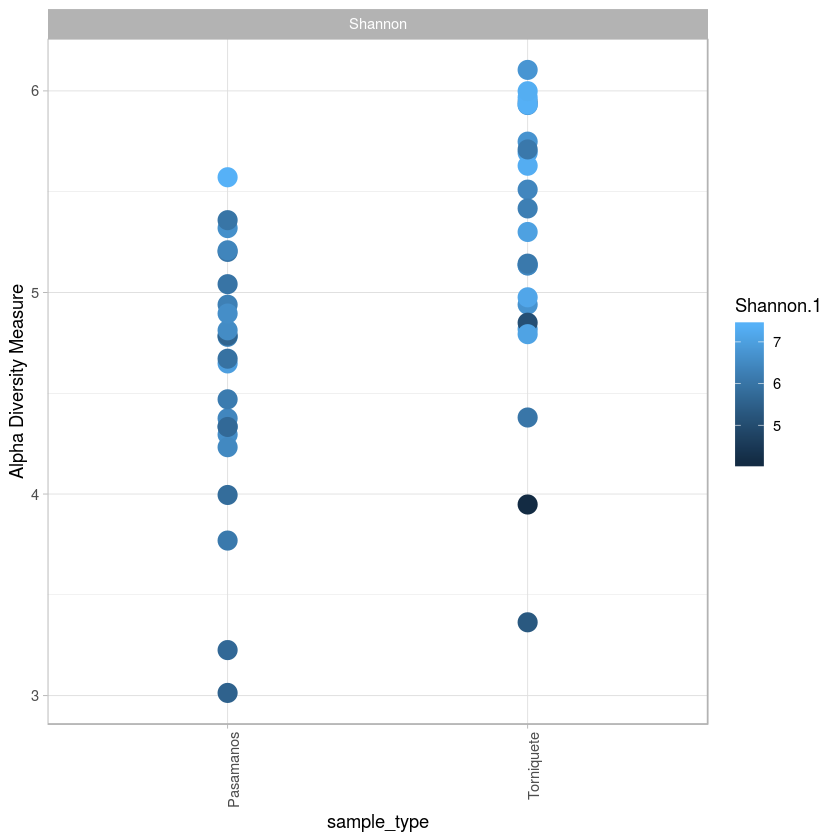
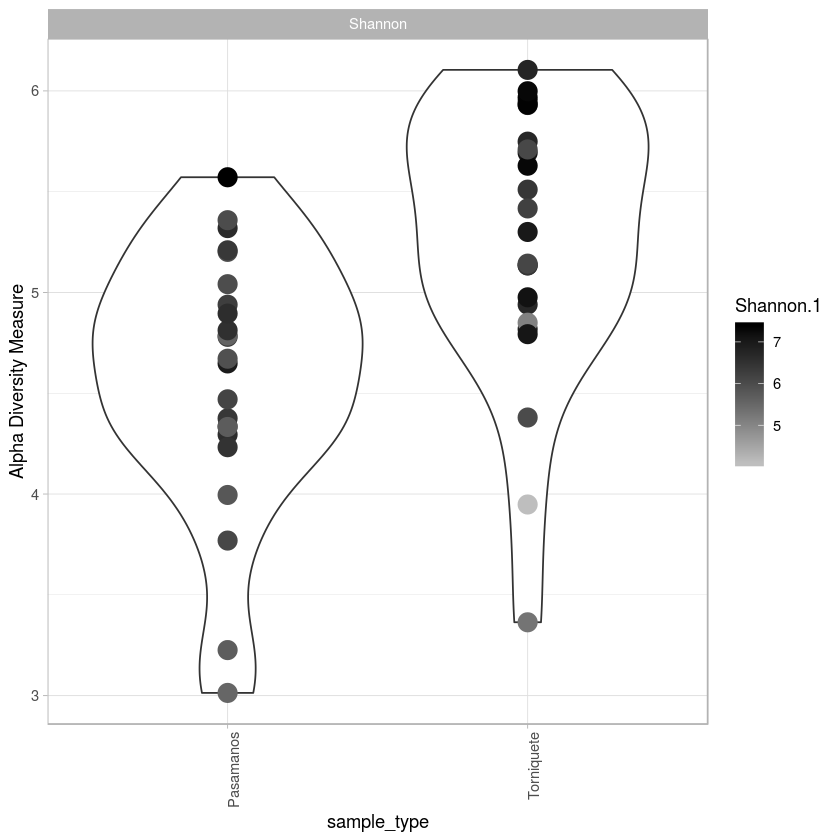
library(ggrepel)
p <- plot_richness(metro, x="sample_type",color="Shannon.1", measures= "Shannon")
p + geom_violin()+ geom_point(aes(colour=a$Shannon.1),size=5, alpha=1) + scale_colour_gradient(low="#bebebe", high="#000000", na.value = "white") + theme_light() + theme(axis.text.x=element_text(angle=90, hjust=1))
library(dplyr)
p <- plot_richness(metro, measures=c("Observed", "Chao1", "Shannon", "Simpson")) + geom_boxplot()+theme_light() + theme(axis.text.x=element_text(angle=90, hjust=1))
#p$data
#a <-p$data
#summary(a)
#write.table(a, "data.csv") Se edita para dejar en el formato id Observed Chao1 Shannon Simpson
meta <- read.table("THEmetadata.txt",sep="\t",header=T,row.names = 1) #data.csv se hizo a partir de la tabla anterior para calcular
#los datos de diversidad alfa
head(meta)
summary(meta)
hist(meta$Observed, breaks=10)
hist(meta$Chao1, breaks=10)
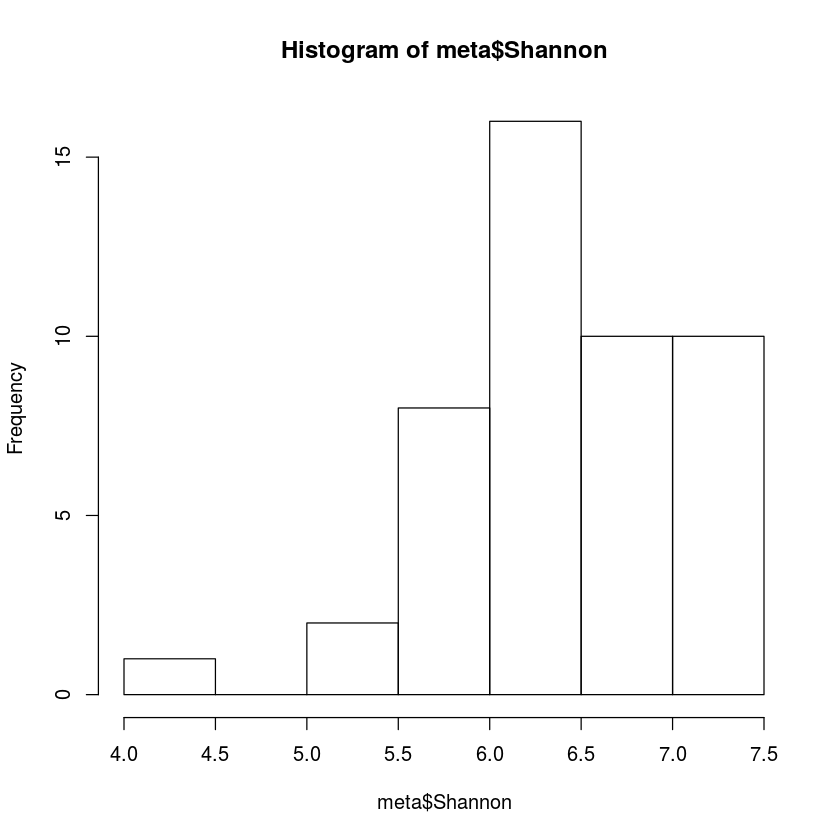
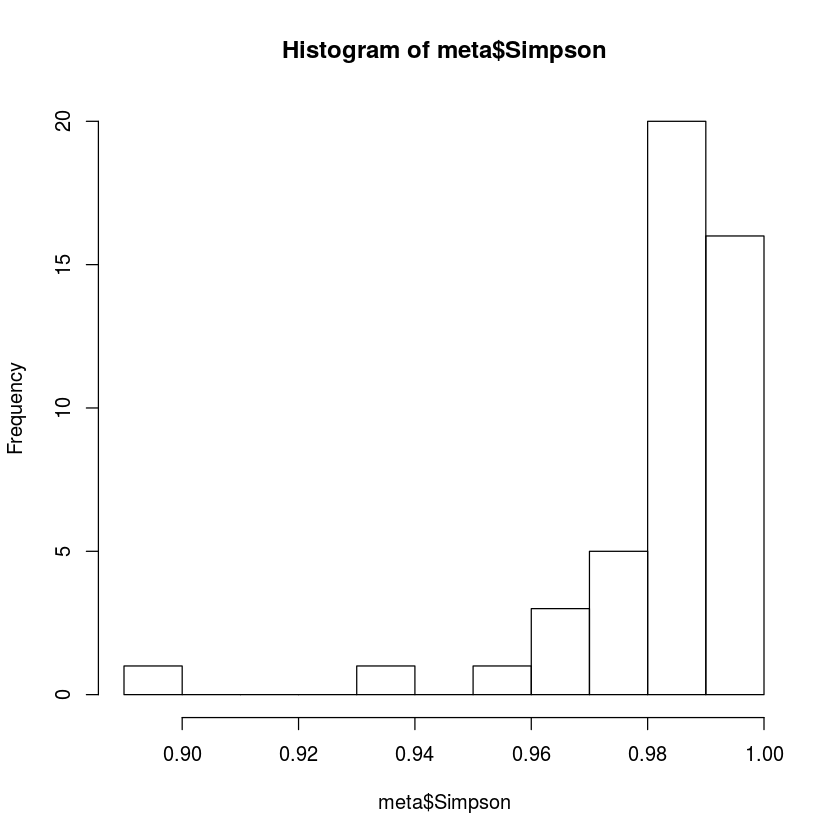
hist(meta$Shannon, breaks=10)
hist(meta$Simpson, breaks=10)
| <th scope=col>otu</th><th scope=col>id</th><th scope=col>sample_location</th><th scope=col>sample_id</th><th scope=col>line_number</th><th scope=col>sample_type</th><th scope=col>date</th><th scope=col>time</th><th scope=col>temperature_C</th><th scope=col>Humidity</th><th scope=col>⋯</th><th scope=col>geographical_zone</th><th scope=col>weather</th><th scope=col>stations_number</th><th scope=col>latitude</th><th scope=col>longitude</th><th scope=col>people_affluence</th><th scope=col>Observed</th><th scope=col>Chao1</th><th scope=col>Shannon</th><th scope=col>Simpson</th> | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Indios_verdes_3 | Indios_verdes_3 | Indios_verdes | AM01 | 3 | Torniquete | 2/5/16 | 2:23:00 PM | 30.4 | 27 | ⋯ | Norte | C1 | 21 | 19.4955 | -99.11940 | 10176457 | 12677 | 19699.2193 | 7.160039 | 0.9921556 |
| Instituto_del_petroleo_5 | Instituto_del_petroleo_5 | Instituto_del_petroleo | AM02 | 5 | Torniquete | 4/16/29 | 1:17:00 PM | 28.3 | 30 | ⋯ | Norte | C1 | 13 | 19.4890 | -99.14480 | 499350 | 10722 | 16496.3811 | 6.417885 | 0.9890120 |
| Rosario_7 | Rosario_7 | Rosario | AM03 | 7 | Torniquete | 4/16/29 | 12:40:00 PM | 29.2 | 29 | ⋯ | Norte | C1 | 14 | 19.5050 | -99.17970 | 3220719 | 11986 | 18521.1304 | 6.718174 | 0.9892510 |
| Cuatro_caminos_2 | Cuatro_caminos_2 | Cuatro_caminos | AM04 | 2 | Torniquete | 4/16/29 | 11:42:00 AM | 30.3 | 30 | ⋯ | Poniente | C2 | 24 | 19.4606 | -99.21550 | 9523016 | 5425 | 8341.2612 | 6.283001 | 0.9939737 |
| Tacubaya_9 | Tacubaya_9 | Tacubaya | AM05 | 9 | Torniquete | 4/16/27 | 1:41:00 PM | 30.4 | 26 | ⋯ | Poniente | C1 | 12 | 19.4024 | -99.18830 | 4190568 | 428 | 840.2462 | 5.309640 | 0.9899266 |
| Observatorio_1 | Observatorio_1 | Observatorio | AM06 | 1 | Torniquete | 4/5/16 | 2:35:00 PM | 32.2 | 28 | ⋯ | Poniente | C2 | 20 | 19.3985 | -99.19978 | 6489055 | 10523 | 17035.5115 | 6.437592 | 0.9859032 |
otu id
Barranca_del_muerto_7 : 1 Barranca_del_muerto_7 : 1
Buenavista_B : 1 Buenavista_B : 1
Chilpancingo_9 : 1 Chilpancingo_9 : 1
Ciudad_Azteca_B : 1 Ciudad_Azteca_B : 1
Constitucion_de_1917_8: 1 Constitucion_de_1917_8: 1
Cuatro_caminos_2 : 1 Cuatro_caminos_2 : 1
(Other) :41 (Other) :41
sample_location sample_id line_number sample_type
Train :23 AM01 : 1 2 : 6 Pasamanos :23
Barranca_del_muerto : 1 AM02 : 1 11(B) : 5 Torniquete:24
Buenavista : 1 AM03 : 1 3 : 5
Chilpancingo : 1 AM04 : 1 1 : 4
Ciudad_Azteca : 1 AM05 : 1 10(A) : 4
Constitucion_de_1917: 1 AM06 : 1 7 : 4
(Other) :19 (Other):41 (Other):19
date time temperature_C Humidity
2/5/16 : 9 11:25:00 AM: 2 Min. :25.90 Min. :25.00
3/5/16 : 7 11:42:00 AM: 2 1st Qu.:29.10 1st Qu.:27.00
4/16/27: 9 1:20:00 PM : 2 Median :30.40 Median :30.00
4/16/28: 6 12:05:00 PM: 2 Mean :30.15 Mean :30.38
4/16/29:10 12:40:00 PM: 2 3rd Qu.:31.30 3rd Qu.:32.00
4/5/16 : 6 12:50:00 PM: 2 Max. :33.30 Max. :41.00
(Other) :35
length_underground length_superficial length_elevated length_total
Min. : 0.00 Min. : 0.000 Min. : 0.000 Min. :11.00
1st Qu.: 5.38 1st Qu.: 0.000 1st Qu.: 0.000 1st Qu.:16.50
Median :11.86 Median : 4.449 Median : 0.000 Median :18.00
Mean :11.05 Mean : 5.485 Mean : 2.001 Mean :18.83
3rd Qu.:16.79 3rd Qu.:10.090 3rd Qu.: 4.185 3rd Qu.:22.00
Max. :18.14 Max. :15.151 Max. :11.533 Max. :25.00
elevation station_hubs train_track
Min. :2231 Correspondecia : 3 Ferreo : 7
1st Qu.:2237 Paso : 6 Neumatico:40
Median :2242 Terminal :11
Mean :2250 Terminal_correspondencia: 4
3rd Qu.:2256 Train :23
Max. :2303
NA's :23
train_notes station_notes geographical_zone weather
Independiente:34 Cajon_subterraneo:11 Centro : 5 B1 : 4
Oruga :13 Elevada : 9 Norte : 7 C1 :16
Superficial : 3 Oriente : 5 C2 : 4
Tunel : 1 Poniente: 3 NA's:23
NA's :23 Sur : 4
NA's :23
stations_number latitude longitude people_affluence
Min. :10.00 Min. :19.29 Min. :-99.22 Min. : 499350
1st Qu.:12.00 1st Qu.:19.39 1st Qu.:-99.17 1st Qu.: 4284103
Median :19.00 Median :19.43 Median :-99.14 Median : 9523016
Mean :17.15 Mean :19.43 Mean :-99.13 Mean :17859153
3rd Qu.:21.00 3rd Qu.:19.49 3rd Qu.:-99.10 3rd Qu.:24196452
Max. :24.00 Max. :19.53 Max. :-98.96 Max. :66232325
NA's :23 NA's :23
Observed Chao1 Shannon Simpson
Min. : 428 Min. : 840.2 Min. :4.102 Min. :0.8984
1st Qu.: 7732 1st Qu.:12826.6 1st Qu.:6.014 1st Qu.:0.9813
Median :10129 Median :15375.8 Median :6.410 Median :0.9872
Mean : 9904 Mean :15141.1 Mean :6.380 Mean :0.9827
3rd Qu.:12332 3rd Qu.:18371.5 3rd Qu.:6.810 3rd Qu.:0.9915
Max. :16179 Max. :23052.1 Max. :7.384 Max. :0.9960
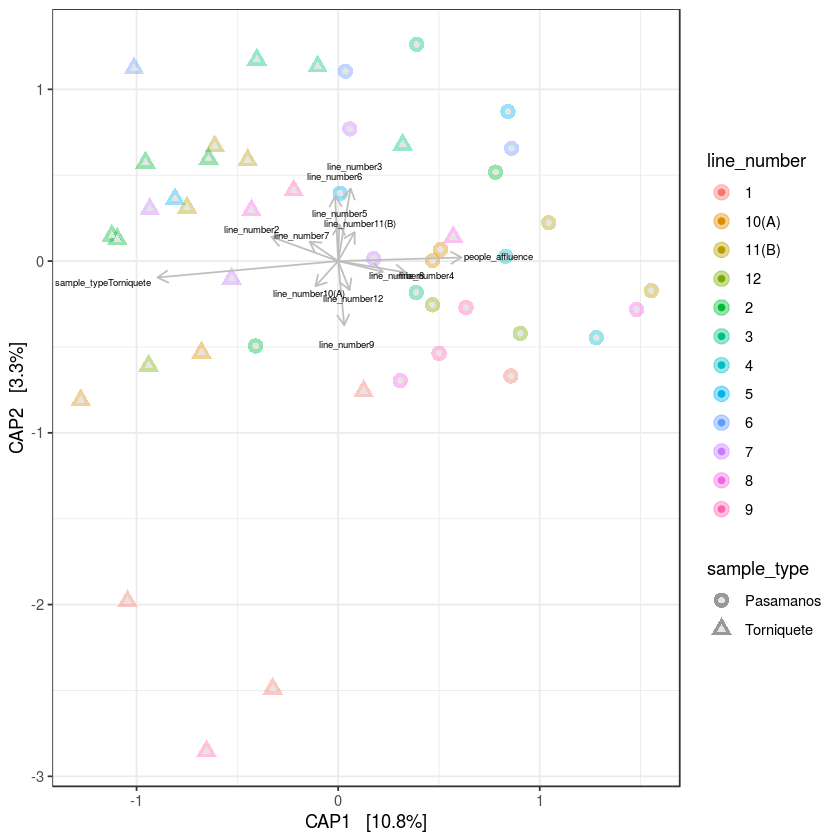
metroCAP.ORD <- ordinate(metro, "CAP", "bray", ~ people_affluence + line_number + sample_type)
metroCAP.ORD
cap_plot <- plot_ordination(metro, metroCAP.ORD, color="line_number", axes =c(1,2)) + aes(shape= sample_type) + geom_point(aes(colour = line_number), alpha=0.4, size=4) + geom_point(colour = "grey90", size=1.5) + theme_bw()
arrowmat <- vegan::scores(metroCAP.ORD, display="bp")
arrowdf <- data.frame(labels=rownames(arrowmat), arrowmat)
# Define the arrow aesthetic mapping
arrow_map <- aes(xend = CAP1,
yend = CAP2,
x = 0,
y = 0,
shape = NULL,
color = NULL,
label = labels)
label_map <- aes(x = 1.3 * CAP1,
y = 1.3 * CAP2,
shape = NULL,
color = NULL,
label = labels)
arrowhead = arrow(length = unit(0.02, "npc"))
cap_plot +
geom_segment(
mapping = arrow_map,
size = .5,
data = arrowdf,
color = "gray",
arrow = arrowhead
) +
geom_text(
mapping = label_map,
size = 2,
data = arrowdf,
show.legend = FALSE
)
an <- anova(metroCAP.ORD, permutations=9999)
an
Call: capscale(formula = OTU ~ people_affluence + line_number +
sample_type, data = data, distance = distance)
Inertia Proportion Rank
Total 9.2927 1.0000
Constrained 3.0265 0.3257 13
Unconstrained 6.2662 0.6743 33
Inertia is squared Bray distance
Species scores projected from ‘OTU’
Eigenvalues for constrained axes:
CAP1 CAP2 CAP3 CAP4 CAP5 CAP6 CAP7 CAP8 CAP9 CAP10 CAP11
1.0012 0.3099 0.2971 0.2532 0.2313 0.1594 0.1558 0.1498 0.1142 0.1053 0.1002
CAP12 CAP13
0.0776 0.0716
Eigenvalues for unconstrained axes:
MDS1 MDS2 MDS3 MDS4 MDS5 MDS6 MDS7 MDS8
0.7839 0.6623 0.5300 0.4344 0.3182 0.3003 0.2666 0.2429
(Showing 8 of 33 unconstrained eigenvalues)
Warning message:
“Ignoring unknown aesthetics: label”
| <th scope=col>Df</th><th scope=col>SumOfSqs</th><th scope=col>F</th><th scope=col>Pr(>F)</th> | |||
|---|---|---|---|
| 13 | 3.026498 | 1.226055 | 0.0074 |
| 33 | 6.266154 | NA | NA |
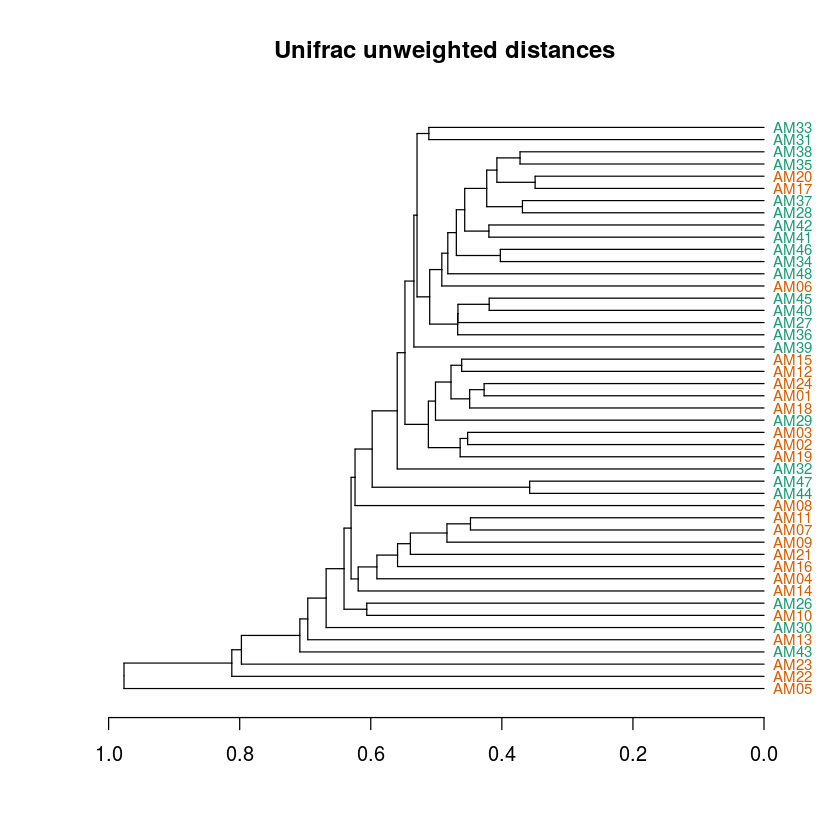
metro
metrito <- get_variable(metro, "sample_type")
sample_data(metro)$metrito <- factor(metrito)
colorCodes <- levels(metrito)
library(doParallel)
library(RColorBrewer)
#sample_data(march_subset)
# camaronUF <- dist(metro, "bray")
camaronUF <- distance(metro, method='bray')
#colorScale <- colors()[c(26,51,76)]
#colorScale <-rainbow(length(levels(get_variable(march_subset, "marchantita"))))
colorScale <- brewer.pal(length(levels(get_variable(metro, "metrito"))),"Dark2")
#colorScale
cols <- colorScale[get_variable(metro, "metrito")]
#cols
#march.tip.labels <- as(get_variable(march_subset, "marchantita"), "character")
# This is the actual hierarchical clustering call, specifying average-link clustering
camaron.hclust <- hclust(camaronUF, method="average")
library(dendextend)
dend <- as.dendrogram(camaron.hclust)
labels_colors(dend) <- cols[order.dendrogram(dend)]
dend %>% set("labels_cex", .75) %>% plot(main="Unifrac unweighted distances",horiz=T)
#ggplot(dend, horiz=T)
phyloseq-class experiment-level object
otu_table() OTU Table: [ 22673 taxa and 47 samples ]
sample_data() Sample Data: [ 47 samples by 29 sample variables ]
tax_table() Taxonomy Table: [ 22673 taxa by 6 taxonomic ranks ]
Warning message in brewer.pal(length(levels(get_variable(metro, "metrito"))), "Dark2"):
“minimal value for n is 3, returning requested palette with 3 different levels
”
---------------------
Welcome to dendextend version 1.10.0
Type citation('dendextend') for how to cite the package.
Type browseVignettes(package = 'dendextend') for the package vignette.
The github page is: https://github.com/talgalili/dendextend/
Suggestions and bug-reports can be submitted at: https://github.com/talgalili/dendextend/issues
Or contact: <tal.galili@gmail.com>
To suppress this message use: suppressPackageStartupMessages(library(dendextend))
---------------------
Attaching package: ‘dendextend’
The following objects are masked from ‘package:ape’:
ladderize, rotate
The following object is masked from ‘package:permute’:
shuffle
The following object is masked from ‘package:stats’:
cutree
library(UpSetR)
library(grid)
merge2 = merge_samples(metro, "line_number")
merge2 <- as.table(t(otu_table(merge2)))
merge2 <- replace(merge2, merge2>0, 1)
write.table(merge2,"merge2.tmp")
merge2 <- read.table("merge2.tmp", header=TRUE, row.names = 1)
head(merge2)
#merge2[order(rowSums(merge2),decreasing=T),] #
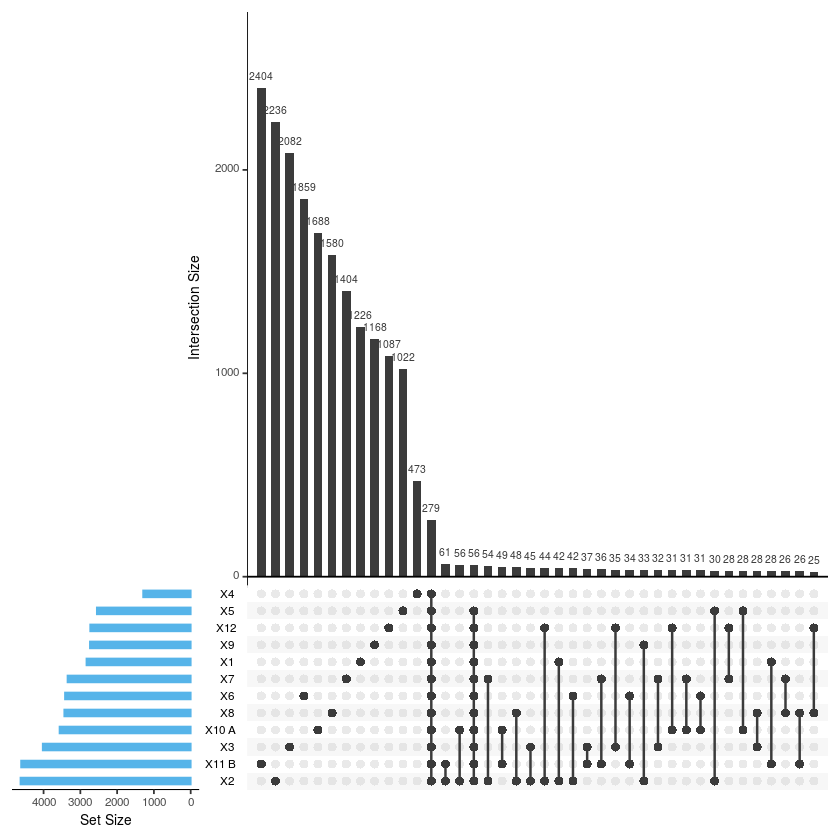
upset(merge2, sets = c('X1','X2','X3','X4','X5','X6','X7','X8','X9','X10.A.','X11.B.', 'X12'),
sets.bar.color = "#56B4E9", order.by = "freq", empty.intersections = "on")
| <th scope=col>X1</th><th scope=col>X10.A.</th><th scope=col>X11.B.</th><th scope=col>X12</th><th scope=col>X2</th><th scope=col>X3</th><th scope=col>X4</th><th scope=col>X5</th><th scope=col>X6</th><th scope=col>X7</th><th scope=col>X8</th><th scope=col>X9</th> | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
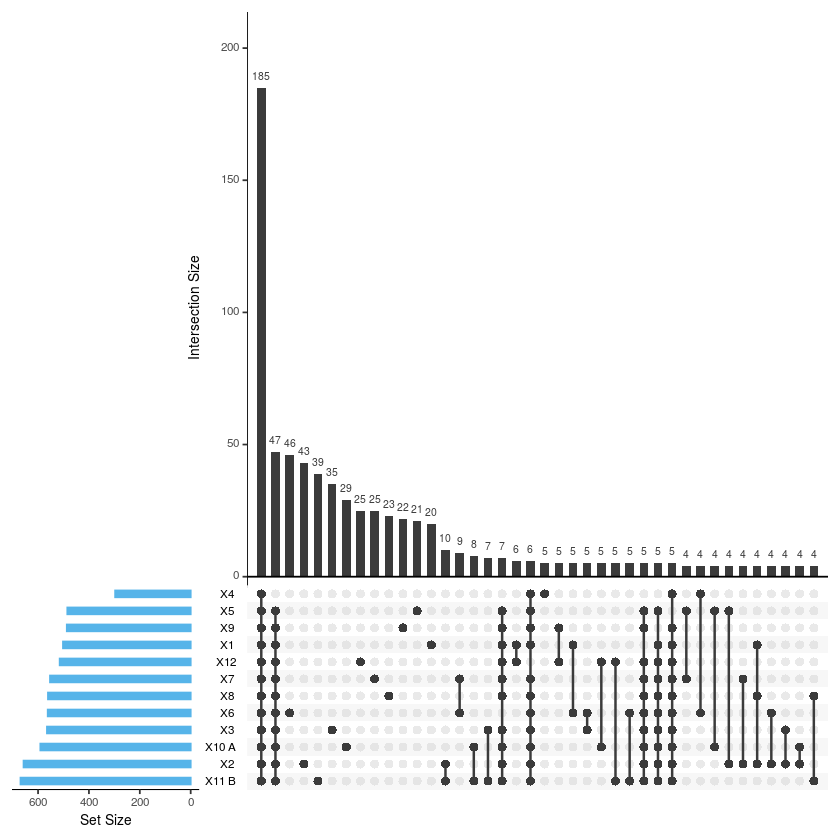
x3 <- tax_glom(metro, taxrank="Genus")
merge2 = merge_samples(x3, "line_number")
merge2 <- as.table(t(otu_table(merge2)))
merge2 <- replace(merge2, merge2>0, 1)
write.table(merge2,"merge2.tmp")
merge2 <- read.table("merge2.tmp", header=TRUE, row.names = 1)
head(merge2)
#merge2[order(rowSums(merge2),decreasing=T),] #
upset(merge2, sets = c('X1','X2','X3','X4','X5','X6','X7','X8','X9','X10.A.','X11.B.', 'X12'),
sets.bar.color = "#56B4E9", order.by = "freq", empty.intersections = "on")
upset(merge2, sets = c('X1','X2','X3','X4','X5','X6','X7','X8','X9','X10.A.','X11.B.', 'X12'),
sets.bar.color = "#56B4E9", order.by = "freq", empty.intersections = "on")
ggsave("upset_lines.pdf", width= 10, height= 10, onefile=FALSE)
| <th scope=col>X1</th><th scope=col>X10.A.</th><th scope=col>X11.B.</th><th scope=col>X12</th><th scope=col>X2</th><th scope=col>X3</th><th scope=col>X4</th><th scope=col>X5</th><th scope=col>X6</th><th scope=col>X7</th><th scope=col>X8</th><th scope=col>X9</th> | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
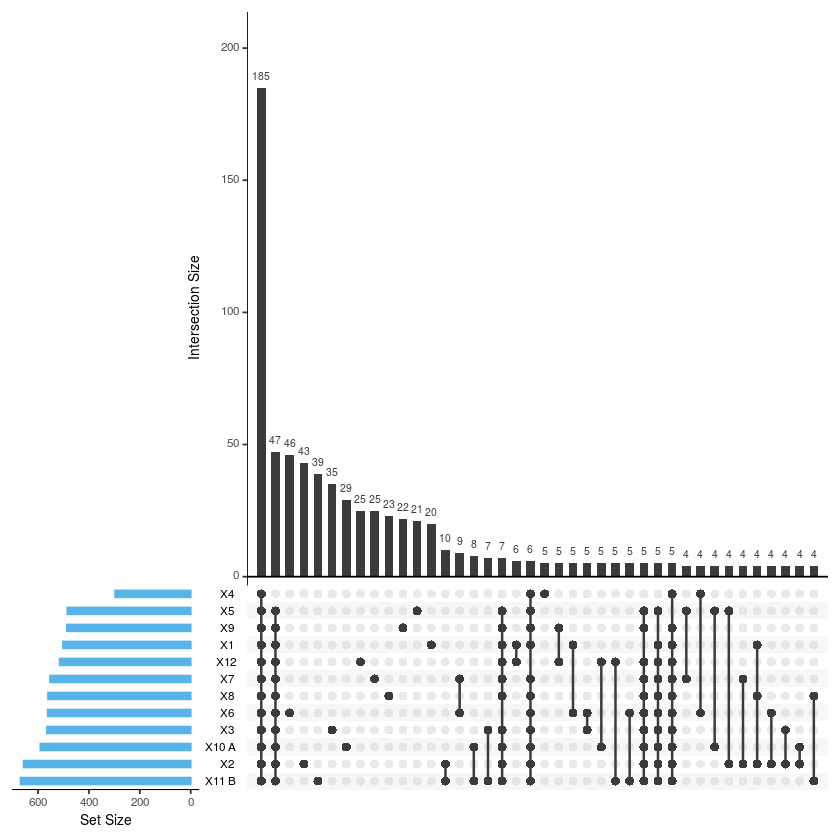
#Función para calcular las intersecciones get_intersect_members
get_intersect_members <- function (x, ...){
require(dplyr)
require(tibble)
x <- x[,sapply(x, is.numeric)][,0<=colMeans(x[,sapply(x, is.numeric)],na.rm=T) & colMeans(x[,sapply(x, is.numeric)],na.rm=T)<=1]
n <- names(x)
x %>% rownames_to_column() -> x
l <- c(...)
a <- intersect(names(x), l)
ar <- vector('list',length(n)+1)
ar[[1]] <- x
i=2
for (item in n) {
if (item %in% a){
if (class(x[[item]])=='integer'){
ar[[i]] <- paste(item, '>= 1')
i <- i + 1
}
} else {
if (class(x[[item]])=='integer'){
ar[[i]] <- paste(item, '== 0')
i <- i + 1
}
}
}
do.call(filter_, ar) %>% column_to_rownames() -> x
return(x)
}
intersect <- get_intersect_members(merge2,c('X1','X2','X3','X4','X5','X6','X7','X8','X9','X10.A.','X11.B.', 'X12')) #intersección del core, se puede cambiar para cualquier subconjunto de intersecciones!
length(row.names(intersect)) #número de elementos del core
intersect <- (row.names(intersect)) #identidades del core
vec <- setNames(nm=c(intersect))
# vec
185
physeqsubsetOTU <- subset_taxa(x3, rownames(tax_table(x3)) %in% vec)
mergeph1 <- merge_samples(physeqsubsetOTU, "line_number")
mergeph1 = transform_sample_counts(mergeph1,function(x) x/sum(x))
write.table(t(otu_table(mergeph1)),"heatmap_core-lineas.csv",sep="\t")
write.table(tax_table(mergeph1),"heatmap_core-lineas.tax.csv",sep="\t")
system("paste heatmap_core-lineas.csv heatmap_core-lineas.tax.csv >heatmap_core-lineas.otutax.csv" )
t1 <- as.data.frame(colSums(otu_table(mergeph1)))
# write.table(t, "vennupset.txt")
t <-(tax_table(mergeph1)) #hago esto porque el melt sobre este objeto phyloseq no trabaja, creo que hay que usar psmelt para esto
write.table(t, "taxvennupset.txt")
t <- as.data.frame(read.table("taxvennupset.txt", header=T, row.names = 1))
# system("paste vennupset.txt taxvennupset.txt >coreupset.txt")
# read.table("coreupset.txt", header=T, sep="\t")
t3 <- merge(t,t1, by=0, all=TRUE )
head(t3)
names(t3)[8] <-"abundance"
t4 <- t3[order(-t3$abundance),]
write.table(t4, "taxvennupset-Entrada-Salida.txt")
t5 <- as.vector(t4$Row.names)
t5
t4
summary(t4)
p <- plot_bar(mergeph1, "Genus")
yx <- p$data
yx <- as.data.frame(yx)
head(yx)
yx <- yx[order(-yx$Abundance),]
head(yx)
levels1=c('1','2','3','4','5','6','7','8','9','10(A)','11(B)','12')
levels1
levels2 = rev(t5)
yx$orden <- factor(yx$Sample, levels = levels1)
yx$abun <- factor(yx$OTU, levels= levels2)
head(yx$abun)
#Correción a valores vacios en Género
ylabvec = as(tax_table(mergeph1)[,"Genus"], "character")
names(ylabvec) <- taxa_names(mergeph1)
ylabvec[is.na(ylabvec)] <- ""
#####
# ggplot(data=yx, aes(x = yx$orden, y= reorder(Genus, Abundance), fill= Abundance)) +
# geom_raster() + theme_minimal () +
# xlab("") + ylab("") +
# scale_fill_gradient(low="#bebebe", high="#000000", na.value = "white", trans = "log10") +
# theme(axis.text.x = element_text( angle = 90, hjust = 1),
# axis.text.y = element_text(size = 4.2))
| ASV_1 | Bacteria | Actinobacteriota | Actinobacteria | Propionibacteriales | Propionibacteriaceae | Cutibacterium | 2.290667670 |
| ASV_1007 | Bacteria | Firmicutes | Clostridia | Clostridia_or | Hungateiclostridiaceae | Fastidiosipila | 0.001820265 |
| ASV_101 | Bacteria | Proteobacteria | Alphaproteobacteria | Azospirillales | Azospirillaceae | Skermanella | 0.020332544 |
| ASV_1027 | Bacteria | Firmicutes | Clostridia | Peptostreptococcales-Tissierellales | Peptostreptococcales-Tissierellales_fa | Murdochiella | 0.001554152 |
| ASV_1039 | Bacteria | Bacteroidota | Bacteroidia | Cytophagales | Hymenobacteraceae | Adhaeribacter | 0.005039240 |
| ASV_1044 | Bacteria | Firmicutes | Clostridia | Lachnospirales | Lachnospiraceae | Lachnoclostridium | 0.001689947 |
<ol class=list-inline> <li>‘ASV_1’</li> <li>‘ASV_2’</li> <li>‘ASV_6’</li> <li>‘ASV_5’</li> <li>‘ASV_4’</li> <li>‘ASV_3’</li> <li>‘ASV_13’</li> <li>‘ASV_26’</li> <li>‘ASV_19’</li> <li>‘ASV_9’</li> <li>‘ASV_70’</li> <li>‘ASV_18’</li> <li>‘ASV_90’</li> <li>‘ASV_96’</li> <li>‘ASV_17’</li> <li>‘ASV_16’</li> <li>‘ASV_43’</li> <li>‘ASV_28’</li> <li>‘ASV_14’</li> <li>‘ASV_304’</li> <li>‘ASV_31’</li> <li>‘ASV_58’</li> <li>‘ASV_129’</li> <li>‘ASV_42’</li> <li>‘ASV_35’</li> <li>‘ASV_27’</li> <li>‘ASV_41’</li> <li>‘ASV_123’</li> <li>‘ASV_32’</li> <li>‘ASV_82’</li> <li>‘ASV_91’</li> <li>‘ASV_251’</li> <li>‘ASV_66’</li> <li>‘ASV_56’</li> <li>‘ASV_62’</li> <li>‘ASV_184’</li> <li>‘ASV_171’</li> <li>‘ASV_168’</li> <li>‘ASV_95’</li> <li>‘ASV_128’</li> <li>‘ASV_109’</li> <li>‘ASV_36’</li> <li>‘ASV_134’</li> <li>‘ASV_46’</li> <li>‘ASV_52’</li> <li>‘ASV_206’</li> <li>‘ASV_60’</li> <li>‘ASV_54’</li> <li>‘ASV_270’</li> <li>‘ASV_265’</li> <li>‘ASV_412’</li> <li>‘ASV_106’</li> <li>‘ASV_68’</li> <li>‘ASV_77’</li> <li>‘ASV_97’</li> <li>‘ASV_202’</li> <li>‘ASV_137’</li> <li>‘ASV_455’</li> <li>‘ASV_256’</li> <li>‘ASV_74’</li> <li>‘ASV_317’</li> <li>‘ASV_67’</li> <li>‘ASV_117’</li> <li>‘ASV_126’</li> <li>‘ASV_150’</li> <li>‘ASV_118’</li> <li>‘ASV_110’</li> <li>‘ASV_471’</li> <li>‘ASV_253’</li> <li>‘ASV_101’</li> <li>‘ASV_238’</li> <li>‘ASV_447’</li> <li>‘ASV_360’</li> <li>‘ASV_259’</li> <li>‘ASV_125’</li> <li>‘ASV_193’</li> <li>‘ASV_541’</li> <li>‘ASV_397’</li> <li>‘ASV_170’</li> <li>‘ASV_245’</li> <li>‘ASV_138’</li> <li>‘ASV_450’</li> <li>‘ASV_112’</li> <li>‘ASV_136’</li> <li>‘ASV_151’</li> <li>‘ASV_342’</li> <li>‘ASV_274’</li> <li>‘ASV_194’</li> <li>‘ASV_485’</li> <li>‘ASV_431’</li> <li>‘ASV_469’</li> <li>‘ASV_220’</li> <li>‘ASV_543’</li> <li>‘ASV_367’</li> <li>‘ASV_178’</li> <li>‘ASV_1064’</li> <li>‘ASV_215’</li> <li>‘ASV_681’</li> <li>‘ASV_153’</li> <li>‘ASV_510’</li> <li>‘ASV_320’</li> <li>‘ASV_285’</li> <li>‘ASV_191’</li> <li>‘ASV_433’</li> <li>‘ASV_651’</li> <li>‘ASV_214’</li> <li>‘ASV_372’</li> <li>‘ASV_250’</li> <li>‘ASV_174’</li> <li>‘ASV_388’</li> <li>‘ASV_482’</li> <li>‘ASV_357’</li> <li>‘ASV_2015’</li> <li>‘ASV_188’</li> <li>‘ASV_204’</li> <li>‘ASV_226’</li> <li>‘ASV_219’</li> <li>‘ASV_849’</li> <li>‘ASV_319’</li> <li>‘ASV_519’</li> <li>‘ASV_355’</li> <li>‘ASV_440’</li> <li>‘ASV_407’</li> <li>‘ASV_449’</li> <li>‘ASV_901’</li> <li>‘ASV_435’</li> <li>‘ASV_386’</li> <li>‘ASV_553’</li> <li>‘ASV_418’</li> <li>‘ASV_815’</li> <li>‘ASV_873’</li> <li>‘ASV_1379’</li> <li>‘ASV_1110’</li> <li>‘ASV_1039’</li> <li>‘ASV_445’</li> <li>‘ASV_678’</li> <li>‘ASV_326’</li> <li>‘ASV_942’</li> <li>‘ASV_3196’</li> <li>‘ASV_375’</li> <li>‘ASV_609’</li> <li>‘ASV_830’</li> <li>‘ASV_590’</li> <li>‘ASV_999’</li> <li>‘ASV_755’</li> <li>‘ASV_684’</li> <li>‘ASV_2142’</li> <li>‘ASV_963’</li> <li>‘ASV_1060’</li> <li>‘ASV_480’</li> <li>‘ASV_1254’</li> <li>‘ASV_525’</li> <li>‘ASV_1623’</li> <li>‘ASV_659’</li> <li>‘ASV_836’</li> <li>‘ASV_2541’</li> <li>‘ASV_1137’</li> <li>‘ASV_935’</li> <li>‘ASV_1869’</li> <li>‘ASV_518’</li> <li>‘ASV_1186’</li> <li>‘ASV_3882’</li> <li>‘ASV_1472’</li> <li>‘ASV_931’</li> <li>‘ASV_2005’</li> <li>‘ASV_1719’</li> <li>‘ASV_1342’</li> <li>‘ASV_2962’</li> <li>‘ASV_1965’</li> <li>‘ASV_1348’</li> <li>‘ASV_1752’</li> <li>‘ASV_2924’</li> <li>‘ASV_1007’</li> <li>‘ASV_3880’</li> <li>‘ASV_1044’</li> <li>‘ASV_1984’</li> <li>‘ASV_1027’</li> <li>‘ASV_4864’</li> <li>‘ASV_1196’</li> <li>‘ASV_2195’</li> <li>‘ASV_5031’</li> <li>‘ASV_1440’</li> <li>‘ASV_4584’</li> <li>‘ASV_3549’</li> <li>‘ASV_1973’</li> </ol>
| <th scope=col>Row.names</th><th scope=col>Kingdom</th><th scope=col>Phylum</th><th scope=col>Class</th><th scope=col>Order</th><th scope=col>Family</th><th scope=col>Genus</th><th scope=col>abundance</th> | |||||||
|---|---|---|---|---|---|---|---|
| ASV_1 | Bacteria | Actinobacteriota | Actinobacteria | Propionibacteriales | Propionibacteriaceae | Cutibacterium | 2.29066767 |
| ASV_2 | Bacteria | Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | Streptococcus | 1.11725948 |
| ASV_6 | Bacteria | Actinobacteriota | Actinobacteria | Corynebacteriales | Corynebacteriaceae | Corynebacterium | 1.06249797 |
| ASV_5 | Bacteria | Firmicutes | Bacilli | Staphylococcales | Staphylococcaceae | Staphylococcus | 0.68366594 |
| ASV_4 | Bacteria | Actinobacteriota | Actinobacteria | Corynebacteriales | Corynebacteriaceae | Lawsonella | 0.53559802 |
| ASV_3 | Bacteria | Proteobacteria | Gammaproteobacteria | Oceanospirillales | Halomonadaceae | Halomonas | 0.40929609 |
| ASV_13 | Bacteria | Actinobacteriota | Actinobacteria | Micrococcales | Micrococcaceae | Rothia | 0.39471905 |
| ASV_26 | Bacteria | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Moraxellaceae | Acinetobacter | 0.34119212 |
| ASV_19 | Bacteria | Actinobacteriota | Actinobacteria | Micrococcales | Micrococcaceae | Kocuria | 0.33427959 |
| ASV_9 | Bacteria | Actinobacteriota | Actinobacteria | Micrococcales | Micrococcaceae | Micrococcus | 0.26203884 |
| ASV_70 | Bacteria | Actinobacteriota | Actinobacteria | Actinomycetales | Actinomycetaceae | Actinomyces | 0.25964354 |
| ASV_18 | Bacteria | Proteobacteria | Alphaproteobacteria | Rhizobiales | Beijerinckiaceae | Methylobacterium-Methylorubrum | 0.18500243 |
| ASV_90 | Bacteria | Bacteroidota | Bacteroidia | Bacteroidales | Prevotellaceae | Prevotella | 0.17138229 |
| ASV_96 | Bacteria | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | Pseudomonas | 0.17136659 |
| ASV_17 | Bacteria | Proteobacteria | Alphaproteobacteria | Rhodobacterales | Rhodobacteraceae | Paracoccus | 0.14373532 |
| ASV_16 | Bacteria | Firmicutes | Bacilli | Lactobacillales | Leuconostocaceae | Leuconostoc | 0.13999839 |
| ASV_43 | Bacteria | Firmicutes | Negativicutes | Veillonellales-Selenomonadales | Veillonellaceae | Veillonella | 0.12991747 |
| ASV_28 | Bacteria | Actinobacteriota | Actinobacteria | Micrococcales | Dermabacteraceae | Brachybacterium | 0.12977183 |
| ASV_14 | Bacteria | Proteobacteria | Gammaproteobacteria | Alteromonadales | Shewanellaceae | Shewanella | 0.12430211 |
| ASV_304 | Bacteria | Actinobacteriota | Actinobacteria | Propionibacteriales | Nocardioidaceae | Nocardioides | 0.09656966 |
| ASV_31 | Bacteria | Firmicutes | Bacilli | Lactobacillales | Leuconostocaceae | Weissella | 0.09483407 |
| ASV_58 | Bacteria | Firmicutes | Clostridia | Peptostreptococcales-Tissierellales | Peptostreptococcales-Tissierellales_fa | Anaerococcus | 0.08978460 |
| ASV_129 | Bacteria | Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | Lactobacillus | 0.08136070 |
| ASV_42 | Bacteria | Proteobacteria | Gammaproteobacteria | Pasteurellales | Pasteurellaceae | Haemophilus | 0.08001474 |
| ASV_35 | Bacteria | Actinobacteriota | Actinobacteria | Frankiales | Geodermatophilaceae | Blastococcus | 0.07522387 |
| ASV_27 | Bacteria | Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | Lactococcus | 0.07123338 |
| ASV_41 | Bacteria | Firmicutes | Bacilli | Staphylococcales | Gemellaceae | Gemella | 0.06659775 |
| ASV_123 | Bacteria | Firmicutes | Clostridia | Clostridiales | Clostridiaceae | Clostridium_sensu_stricto_1 | 0.06305577 |
| ASV_32 | Bacteria | Actinobacteriota | Actinobacteria | Micrococcales | Intrasporangiaceae | Janibacter | 0.06215911 |
| ASV_82 | Bacteria | Proteobacteria | Gammaproteobacteria | Burkholderiales | Neisseriaceae | Neisseria | 0.06157533 |
| ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ |
| ASV_2541 | Bacteria | Patescibacteria | Saccharimonadia | Saccharimonadales | Saccharimonadaceae | TM7a | 0.0033428207 |
| ASV_1137 | Bacteria | Proteobacteria | Alphaproteobacteria | Rhizobiales | Devosiaceae | Devosia | 0.0031977325 |
| ASV_935 | Bacteria | Firmicutes | Bacilli | Bacillales | Planococcaceae | Kurthia | 0.0031532184 |
| ASV_1869 | Bacteria | Proteobacteria | Gammaproteobacteria | Enterobacterales | Yersiniaceae | Serratia | 0.0031374181 |
| ASV_518 | Bacteria | Firmicutes | Bacilli | Lactobacillales | Aerococcaceae | Eremococcus | 0.0027469601 |
| ASV_1186 | Bacteria | Firmicutes | Clostridia | Oscillospirales | Oscillospiraceae | UCG-002 | 0.0026775756 |
| ASV_3882 | Bacteria | Actinobacteriota | Thermoleophilia | Gaiellales | Gaiellaceae | Gaiella | 0.0025280925 |
| ASV_1472 | Bacteria | Firmicutes | Bacilli | Bacillales | Planococcaceae | Lysinibacillus | 0.0025058386 |
| ASV_931 | Bacteria | Firmicutes | Clostridia | Lachnospirales | Lachnospiraceae | Dorea | 0.0025054089 |
| ASV_2005 | Bacteria | Acidobacteriota | Acidobacteriae | Bryobacterales | Bryobacteraceae | Bryobacter | 0.0024479238 |
| ASV_1719 | Bacteria | Proteobacteria | Gammaproteobacteria | Cellvibrionales | Cellvibrionaceae | Cellvibrio | 0.0024149934 |
| ASV_1342 | Bacteria | Firmicutes | Negativicutes | Veillonellales-Selenomonadales | Veillonellaceae | Negativicoccus | 0.0023887852 |
| ASV_2962 | Bacteria | Actinobacteriota | Actinobacteria | Actinomycetales | Actinomycetaceae | Varibaculum | 0.0023235953 |
| ASV_1965 | Bacteria | Synergistota | Synergistia | Synergistales | Synergistaceae | Fretibacterium | 0.0023030874 |
| ASV_1348 | Bacteria | Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Altererythrobacter | 0.0019679164 |
| ASV_1752 | Bacteria | Firmicutes | Clostridia | Peptostreptococcales-Tissierellales | Peptostreptococcales-Tissierellales_fa | W5053 | 0.0019378517 |
| ASV_2924 | Bacteria | Firmicutes | Clostridia | Oscillospirales | Oscillospiraceae | UCG-005 | 0.0019250371 |
| ASV_1007 | Bacteria | Firmicutes | Clostridia | Clostridia_or | Hungateiclostridiaceae | Fastidiosipila | 0.0018202651 |
| ASV_3880 | Bacteria | Gemmatimonadota | Gemmatimonadetes | Gemmatimonadales | Gemmatimonadaceae | Roseisolibacter | 0.0017879066 |
| ASV_1044 | Bacteria | Firmicutes | Clostridia | Lachnospirales | Lachnospiraceae | Lachnoclostridium | 0.0016899467 |
| ASV_1984 | Bacteria | Firmicutes | Clostridia | Peptococcales | Peptococcaceae | Peptococcus | 0.0016149387 |
| ASV_1027 | Bacteria | Firmicutes | Clostridia | Peptostreptococcales-Tissierellales | Peptostreptococcales-Tissierellales_fa | Murdochiella | 0.0015541522 |
| ASV_4864 | Bacteria | Firmicutes | Bacilli | Paenibacillales | Paenibacillaceae | Paenibacillus | 0.0015324177 |
| ASV_1196 | Bacteria | Bacteroidota | Bacteroidia | Bacteroidales | Tannerellaceae | Tannerella | 0.0014439009 |
| ASV_2195 | Bacteria | Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | Floricoccus | 0.0013014153 |
| ASV_5031 | Bacteria | Abditibacteriota | Abditibacteria | Abditibacteriales | Abditibacteriaceae | Abditibacterium | 0.0011572429 |
| ASV_1440 | Bacteria | Proteobacteria | Gammaproteobacteria | Cardiobacteriales | Cardiobacteriaceae | Cardiobacterium | 0.0011498768 |
| ASV_4584 | Bacteria | Bacteroidota | Bacteroidia | Bacteroidales | Rikenellaceae | Rikenellaceae_RC9_gut_group | 0.0011047558 |
| ASV_3549 | Bacteria | Firmicutes | Clostridia | Lachnospirales | Defluviitaleaceae | Defluviitaleaceae_UCG-011 | 0.0005573410 |
| ASV_1973 | Bacteria | Actinobacteriota | Actinobacteria | Micrococcales | Micrococcaceae | Acaricomes | 0.0005485926 |
Row.names Kingdom Phylum
Length:185 Bacteria:185 Firmicutes :70
Class :AsIs Actinobacteriota:50
Mode :character Proteobacteria :40
Bacteroidota :15
Deinococcota : 2
Fusobacteriota : 2
(Other) : 6
Class Order
Actinobacteria :46 Micrococcales : 24
Bacilli :33 Lactobacillales : 18
Clostridia :31 Peptostreptococcales-Tissierellales: 14
Gammaproteobacteria:28 Lachnospirales : 9
Bacteroidia :15 Corynebacteriales : 7
Alphaproteobacteria:12 Staphylococcales : 7
(Other) :20 (Other) :106
Family Genus
Micrococcaceae : 9 Abditibacterium: 1
Lachnospiraceae : 8 Abiotrophia : 1
Peptostreptococcales-Tissierellales_fa: 8 Acaricomes : 1
Carnobacteriaceae : 6 Acidovorax : 1
Staphylococcaceae : 6 Acinetobacter : 1
Peptostreptococcaceae : 5 Actinobacillus : 1
(Other) :143 (Other) :179
abundance
Min. :0.0005486
1st Qu.:0.0045952
Median :0.0131576
Mean :0.0648649
3rd Qu.:0.0379244
Max. :2.2906677
| <th scope=col>OTU</th><th scope=col>Sample</th><th scope=col>Abundance</th><th scope=col>id</th><th scope=col>sample_location</th><th scope=col>sample_id</th><th scope=col>line_number</th><th scope=col>sample_type</th><th scope=col>date</th><th scope=col>time</th><th scope=col>⋯</th><th scope=col>Chao1</th><th scope=col>Shannon</th><th scope=col>Simpson</th><th scope=col>metrito</th><th scope=col>Kingdom</th><th scope=col>Phylum</th><th scope=col>Class</th><th scope=col>Order</th><th scope=col>Family</th><th scope=col>Genus</th> | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ASV_1 | 4 | 0.3753143 | 30.5 | 23.00000 | 33.00000 | 7 | 1.000000 | 1.0 | 21.00000 | ⋯ | 14102.46 | 6.122639 | 0.9801934 | 1.000000 | Bacteria | Actinobacteriota | Actinobacteria | Propionibacteriales | Propionibacteriaceae | Cutibacterium |
| ASV_1 | 8 | 0.2685063 | 25.0 | 17.25000 | 26.50000 | 11 | 1.500000 | 2.5 | 17.75000 | ⋯ | 18272.34 | 6.422750 | 0.9801182 | 1.500000 | Bacteria | Actinobacteriota | Actinobacteria | Propionibacteriales | Propionibacteriaceae | Cutibacterium |
| ASV_1 | 11(B) | 0.2211542 | 21.8 | 13.20000 | 25.00000 | 3 | 1.600000 | 1.6 | 17.60000 | ⋯ | 17894.83 | 6.613091 | 0.9839803 | 1.600000 | Bacteria | Actinobacteriota | Actinobacteria | Propionibacteriales | Propionibacteriaceae | Cutibacterium |
| ASV_1 | 9 | 0.2142901 | 26.0 | 17.25000 | 26.25000 | 12 | 1.500000 | 4.5 | 30.75000 | ⋯ | 12867.00 | 6.131456 | 0.9873997 | 1.500000 | Bacteria | Actinobacteriota | Actinobacteria | Propionibacteriales | Propionibacteriaceae | Cutibacterium |
| ASV_1 | 3 | 0.2041766 | 23.6 | 17.00000 | 18.00000 | 6 | 1.600000 | 2.2 | 19.20000 | ⋯ | 18923.90 | 6.901101 | 0.9882069 | 1.600000 | Bacteria | Actinobacteriota | Actinobacteria | Propionibacteriales | Propionibacteriaceae | Cutibacterium |
| ASV_1 | 12 | 0.1933069 | 23.0 | 22.66667 | 30.66667 | 4 | 1.333333 | 6.0 | 17.33333 | ⋯ | 12046.64 | 6.021041 | 0.9830369 | 1.333333 | Bacteria | Actinobacteriota | Actinobacteria | Propionibacteriales | Propionibacteriaceae | Cutibacterium |
| <th scope=col>OTU</th><th scope=col>Sample</th><th scope=col>Abundance</th><th scope=col>id</th><th scope=col>sample_location</th><th scope=col>sample_id</th><th scope=col>line_number</th><th scope=col>sample_type</th><th scope=col>date</th><th scope=col>time</th><th scope=col>⋯</th><th scope=col>Chao1</th><th scope=col>Shannon</th><th scope=col>Simpson</th><th scope=col>metrito</th><th scope=col>Kingdom</th><th scope=col>Phylum</th><th scope=col>Class</th><th scope=col>Order</th><th scope=col>Family</th><th scope=col>Genus</th> | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ASV_1 | 4 | 0.3753143 | 30.5 | 23.00000 | 33.00000 | 7 | 1.000000 | 1.0 | 21.00000 | ⋯ | 14102.46 | 6.122639 | 0.9801934 | 1.000000 | Bacteria | Actinobacteriota | Actinobacteria | Propionibacteriales | Propionibacteriaceae | Cutibacterium |
| ASV_1 | 8 | 0.2685063 | 25.0 | 17.25000 | 26.50000 | 11 | 1.500000 | 2.5 | 17.75000 | ⋯ | 18272.34 | 6.422750 | 0.9801182 | 1.500000 | Bacteria | Actinobacteriota | Actinobacteria | Propionibacteriales | Propionibacteriaceae | Cutibacterium |
| ASV_1 | 11(B) | 0.2211542 | 21.8 | 13.20000 | 25.00000 | 3 | 1.600000 | 1.6 | 17.60000 | ⋯ | 17894.83 | 6.613091 | 0.9839803 | 1.600000 | Bacteria | Actinobacteriota | Actinobacteria | Propionibacteriales | Propionibacteriaceae | Cutibacterium |
| ASV_1 | 9 | 0.2142901 | 26.0 | 17.25000 | 26.25000 | 12 | 1.500000 | 4.5 | 30.75000 | ⋯ | 12867.00 | 6.131456 | 0.9873997 | 1.500000 | Bacteria | Actinobacteriota | Actinobacteria | Propionibacteriales | Propionibacteriaceae | Cutibacterium |
| ASV_1 | 3 | 0.2041766 | 23.6 | 17.00000 | 18.00000 | 6 | 1.600000 | 2.2 | 19.20000 | ⋯ | 18923.90 | 6.901101 | 0.9882069 | 1.600000 | Bacteria | Actinobacteriota | Actinobacteria | Propionibacteriales | Propionibacteriaceae | Cutibacterium |
| ASV_1 | 12 | 0.1933069 | 23.0 | 22.66667 | 30.66667 | 4 | 1.333333 | 6.0 | 17.33333 | ⋯ | 12046.64 | 6.021041 | 0.9830369 | 1.333333 | Bacteria | Actinobacteriota | Actinobacteria | Propionibacteriales | Propionibacteriaceae | Cutibacterium |
<ol class=list-inline> <li>‘1’</li> <li>‘2’</li> <li>‘3’</li> <li>‘4’</li> <li>‘5’</li> <li>‘6’</li> <li>‘7’</li> <li>‘8’</li> <li>‘9’</li> <li>‘10(A)’</li> <li>‘11(B)’</li> <li>‘12’</li> </ol>
<ol class=list-inline> <li>ASV_1</li> <li>ASV_1</li> <li>ASV_1</li> <li>ASV_1</li> <li>ASV_1</li> <li>ASV_1</li> </ol>
Scale for 'fill' is already present. Adding another scale for 'fill', which
will replace the existing scale.
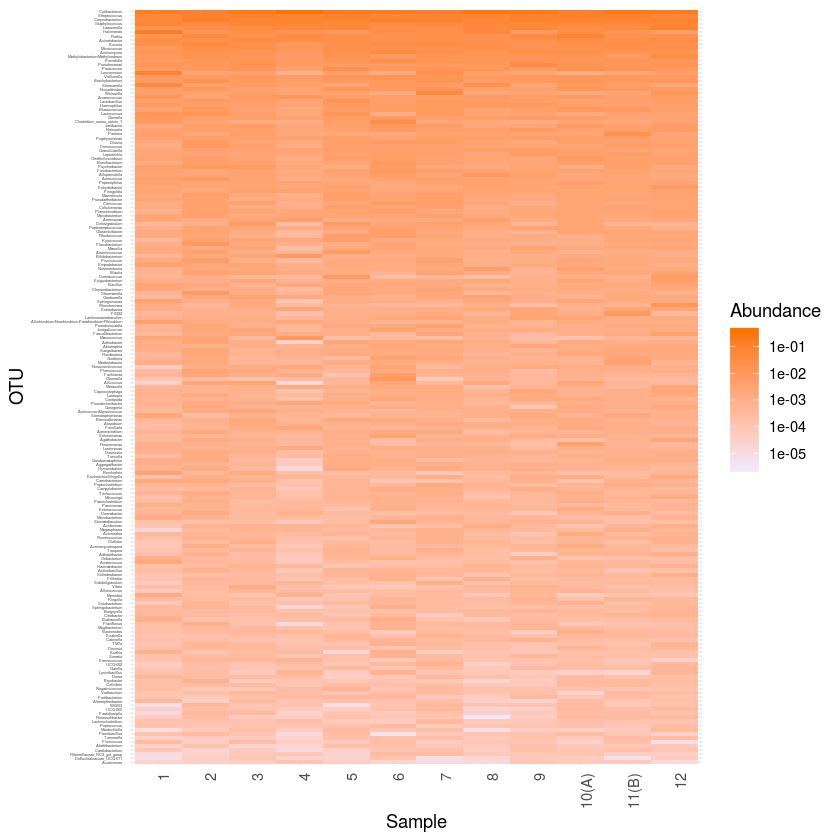
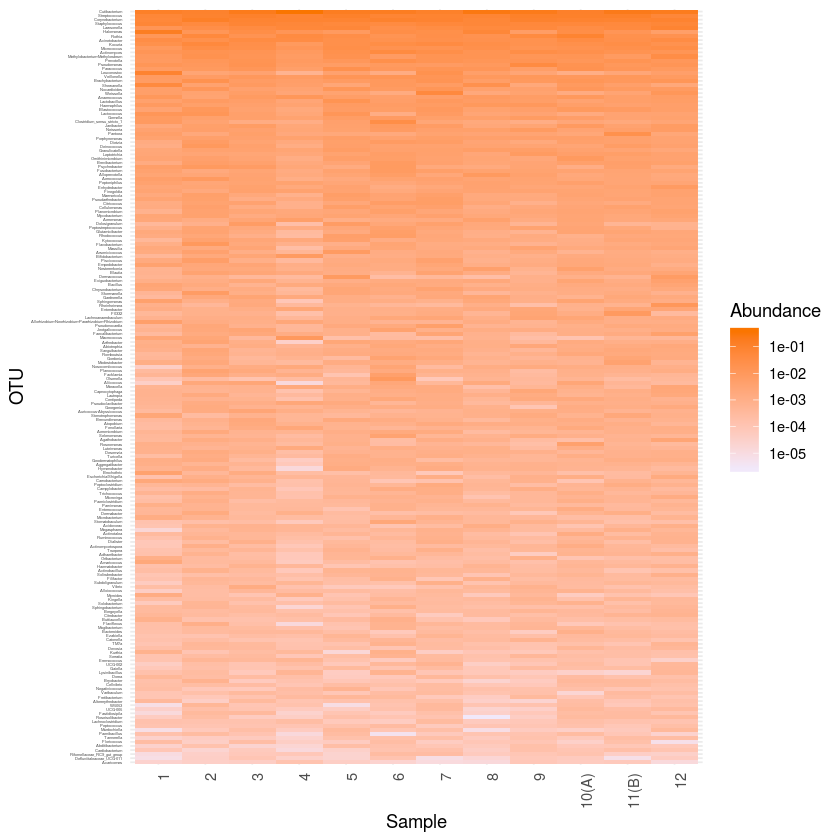
plot_heatmap(mergeph1, "NULL", sample.order=levels1, taxa.order=rev(t5)) + scale_y_discrete(labels=ylabvec) + theme_minimal () +
scale_fill_gradient(low="#F1E8FA", high="#FA7500", na.value = "white", trans = "log10") +
theme(axis.text.x = element_text( angle = 90, hjust = 1),
axis.text.y = element_text(size = 2.5))
ggsave ("heatmap_core.pdf", width=20, height=80, units="cm")
Scale for 'fill' is already present. Adding another scale for 'fill', which
will replace the existing scale.
library(DESeq2)
library(RColorBrewer)
library(ggplot2)
library(phyloseq)
library(plyr)
metro_sig=subset_samples(metro, sample_type !="NA" )
metro_sig
obj1deseq = phyloseq_to_deseq2(metro_sig, ~sample_type)
gm_mean = function(x, na.rm=TRUE){
exp(sum(log(x[x > 0]), na.rm=na.rm) / length(x))
}
geoMeans = apply(counts(obj1deseq), 1, gm_mean)
obj1deseq = estimateSizeFactors(obj1deseq, geoMeans = geoMeans)
##adición de la viñeta para el error de la media de 0s
obj1deseq = DESeq(obj1deseq, fitType="local")
#obj1deseq = DESeq(obj1deseq, test="Wald", fitType="parametric")
Loading required package: S4Vectors
Loading required package: stats4
Loading required package: BiocGenerics
Attaching package: ‘BiocGenerics’
The following objects are masked from ‘package:parallel’:
clusterApply, clusterApplyLB, clusterCall, clusterEvalQ,
clusterExport, clusterMap, parApply, parCapply, parLapply,
parLapplyLB, parRapply, parSapply, parSapplyLB
The following objects are masked from ‘package:dplyr’:
combine, intersect, setdiff, union
The following objects are masked from ‘package:stats’:
IQR, mad, sd, var, xtabs
The following objects are masked from ‘package:base’:
anyDuplicated, append, as.data.frame, basename, cbind, colMeans,
colnames, colSums, dirname, do.call, duplicated, eval, evalq,
Filter, Find, get, grep, grepl, intersect, is.unsorted, lapply,
lengths, Map, mapply, match, mget, order, paste, pmax, pmax.int,
pmin, pmin.int, Position, rank, rbind, Reduce, rowMeans, rownames,
rowSums, sapply, setdiff, sort, table, tapply, union, unique,
unsplit, which, which.max, which.min
Attaching package: ‘S4Vectors’
The following objects are masked from ‘package:dplyr’:
first, rename
The following object is masked from ‘package:tidyr’:
expand
The following object is masked from ‘package:base’:
expand.grid
Loading required package: IRanges
Attaching package: ‘IRanges’
The following object is masked from ‘package:phyloseq’:
distance
The following objects are masked from ‘package:dplyr’:
collapse, desc, slice
The following object is masked from ‘package:purrr’:
reduce
Loading required package: GenomicRanges
Loading required package: GenomeInfoDb
Loading required package: SummarizedExperiment
Loading required package: Biobase
Welcome to Bioconductor
Vignettes contain introductory material; view with
'browseVignettes()'. To cite Bioconductor, see
'citation("Biobase")', and for packages 'citation("pkgname")'.
Attaching package: ‘Biobase’
The following object is masked from ‘package:phyloseq’:
sampleNames
Loading required package: DelayedArray
Loading required package: matrixStats
Attaching package: ‘matrixStats’
The following objects are masked from ‘package:Biobase’:
anyMissing, rowMedians
The following object is masked from ‘package:dplyr’:
count
Loading required package: BiocParallel
Attaching package: ‘DelayedArray’
The following objects are masked from ‘package:matrixStats’:
colMaxs, colMins, colRanges, rowMaxs, rowMins, rowRanges
The following object is masked from ‘package:purrr’:
simplify
The following objects are masked from ‘package:base’:
aperm, apply
------------------------------------------------------------------------------
You have loaded plyr after dplyr - this is likely to cause problems.
If you need functions from both plyr and dplyr, please load plyr first, then dplyr:
library(plyr); library(dplyr)
------------------------------------------------------------------------------
Attaching package: ‘plyr’
The following object is masked from ‘package:matrixStats’:
count
The following object is masked from ‘package:IRanges’:
desc
The following object is masked from ‘package:S4Vectors’:
rename
The following objects are masked from ‘package:dplyr’:
arrange, count, desc, failwith, id, mutate, rename, summarise,
summarize
The following object is masked from ‘package:purrr’:
compact
phyloseq-class experiment-level object
otu_table() OTU Table: [ 22673 taxa and 47 samples ]
sample_data() Sample Data: [ 47 samples by 29 sample variables ]
tax_table() Taxonomy Table: [ 22673 taxa by 6 taxonomic ranks ]
converting counts to integer mode
using pre-existing size factors
estimating dispersions
gene-wise dispersion estimates
mean-dispersion relationship
final dispersion estimates
fitting model and testing
-- replacing outliers and refitting for 8421 genes
-- DESeq argument 'minReplicatesForReplace' = 7
-- original counts are preserved in counts(dds)
estimating dispersions
fitting model and testing
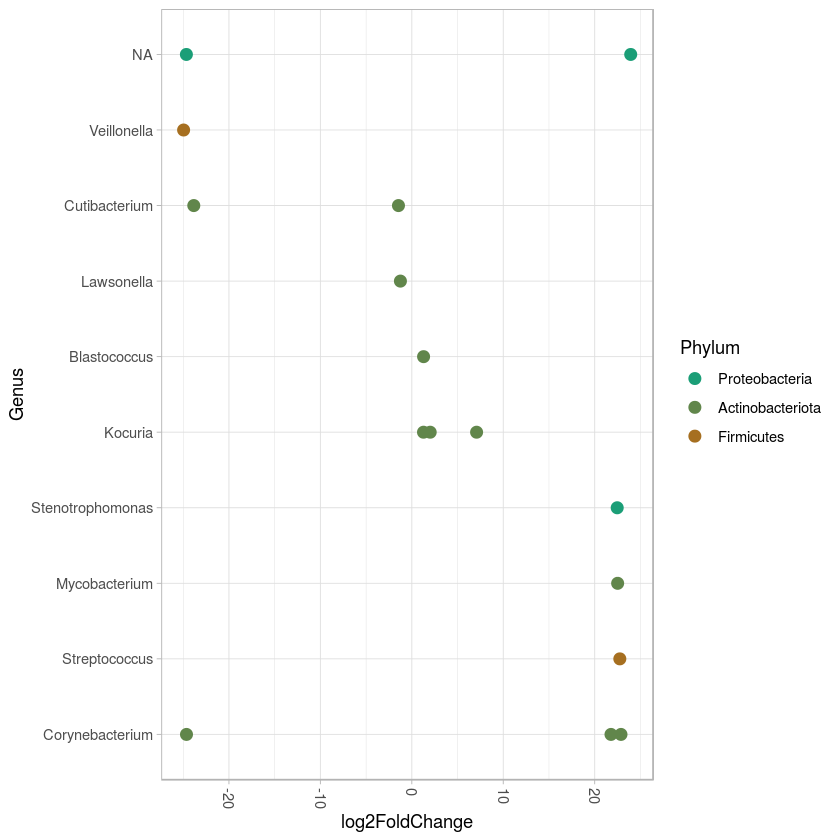
res = results(obj1deseq, cooksCutoff = FALSE)
# quick explanation of Cook's distance: it measures within each gene, for each sample, how removing that sample would change the LFCs (all of the coefficients implied by the design and estimated by DESeq2). So if you have e.g. 3 samples vs 2 samples, and the counts for a gene are [10,10,10] vs [15, 1000], you can see how the Cook's distance will be high for the two samples. Removing either one changes the LFC for the comparison of the two groups. However, if it were [10,10,10] vs [50,50], the two samples "support" each other, such that removing one doesn't change the LFC at all. Hence, we find Cook's to be useful for identifying outliers.
alpha = 0.01
sigtab = res[which(res$padj < alpha), ]
sigtab = cbind(as(sigtab, "data.frame"), as(tax_table(metro_sig)[rownames(sigtab), ], "matrix"))
# Phylum order
x = tapply(sigtab$log2FoldChange, sigtab$Phylum, function(x) max(x))
x = sort(x, TRUE)
sigtab$Phylum = factor(as.character(sigtab$Phylum), levels=names(x))
# Genus order
x = tapply(sigtab$log2FoldChange, sigtab$Genus, function(x) max(x))
x = sort(x, TRUE)
sigtab$Genus = factor(as.character(sigtab$Genus), levels=names(x))
ggplot(sigtab, aes(x=Genus, y=log2FoldChange, color=Phylum)) + scale_color_manual(values=colorRampPalette(brewer.pal(8,"Dark2"))(20))+geom_point(size=3)+theme_light() +theme(axis.text.x = element_text(angle = -90, hjust = 0, vjust=0.5)) + coord_flip()
#sigtab
#ggsave ("torniquete_pasamanos_SIG.pdf", width=30, height=15, units="cm")
sigtab
write.table(sigtab,"handrail-turnstileOR.csv",sep="\t")
| <th scope=col>baseMean</th><th scope=col>log2FoldChange</th><th scope=col>lfcSE</th><th scope=col>stat</th><th scope=col>pvalue</th><th scope=col>padj</th><th scope=col>Kingdom</th><th scope=col>Phylum</th><th scope=col>Class</th><th scope=col>Order</th><th scope=col>Family</th><th scope=col>Genus</th> | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 16132.859322 | -1.459546 | 0.3683595 | -3.962286 | 7.423561e-05 | 6.475629e-03 | Bacteria | Actinobacteriota | Actinobacteria | Propionibacteriales | Propionibacteriaceae | Cutibacterium |
| 2146.727806 | -1.246392 | 0.3191419 | -3.905448 | 9.405103e-05 | 7.110258e-03 | Bacteria | Actinobacteriota | Actinobacteria | Corynebacteriales | Corynebacteriaceae | Lawsonella |
| 497.860423 | 2.009594 | 0.4142819 | 4.850790 | 1.229709e-06 | 1.267718e-04 | Bacteria | Actinobacteriota | Actinobacteria | Micrococcales | Micrococcaceae | Kocuria |
| 469.021045 | 1.278936 | 0.3325598 | 3.845732 | 1.201930e-04 | 8.518676e-03 | Bacteria | Actinobacteriota | Actinobacteria | Micrococcales | Micrococcaceae | Kocuria |
| 286.296527 | 1.286759 | 0.3289152 | 3.912130 | 9.148558e-05 | 7.110258e-03 | Bacteria | Actinobacteriota | Actinobacteria | Frankiales | Geodermatophilaceae | Blastococcus |
| 12.696749 | -24.642029 | 2.9166383 | -8.448778 | 2.943678e-17 | 5.563552e-15 | Bacteria | Actinobacteriota | Actinobacteria | Corynebacteriales | Corynebacteriaceae | Corynebacterium |
| 22.251272 | 23.939615 | 2.0813607 | 11.501906 | 1.290340e-30 | 1.463246e-27 | Bacteria | Proteobacteria | Alphaproteobacteria | Rickettsiales | Mitochondria | NA |
| 7.071797 | -23.844134 | 2.9172184 | -8.173586 | 2.993568e-16 | 4.849580e-14 | Bacteria | Actinobacteriota | Actinobacteria | Propionibacteriales | Propionibacteriaceae | Cutibacterium |
| 10.375836 | 22.875134 | 2.9179937 | 7.839336 | 4.529343e-15 | 6.420344e-13 | Bacteria | Actinobacteriota | Actinobacteria | Corynebacteriales | Corynebacteriaceae | Corynebacterium |
| 16.617408 | 21.782770 | 2.9176957 | 7.465744 | 8.283011e-14 | 9.392935e-12 | Bacteria | Actinobacteriota | Actinobacteria | Corynebacteriales | Corynebacteriaceae | Corynebacterium |
| 7.981757 | 22.510169 | 2.4877131 | 9.048539 | 1.448943e-19 | 3.286202e-17 | Bacteria | Actinobacteriota | Actinobacteria | Corynebacteriales | Mycobacteriaceae | Mycobacterium |
| 14.272812 | 7.084544 | 1.6714053 | 4.238675 | 2.248426e-05 | 2.124763e-03 | Bacteria | Actinobacteriota | Actinobacteria | Micrococcales | Micrococcaceae | Kocuria |
| 16.283553 | -24.957553 | 2.2214287 | -11.234911 | 2.747721e-29 | 1.557958e-26 | Bacteria | Firmicutes | Negativicutes | Veillonellales-Selenomonadales | Veillonellaceae | Veillonella |
| 13.255468 | -24.653074 | 2.5092269 | -9.824968 | 8.790189e-23 | 3.322691e-20 | Bacteria | Proteobacteria | Gammaproteobacteria | Burkholderiales | Neisseriaceae | NA |
| 9.475545 | 22.748061 | 2.5060227 | 9.077356 | 1.112438e-19 | 3.153763e-17 | Bacteria | Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | Streptococcus |
| 7.761658 | 22.461984 | 2.9182616 | 7.697042 | 1.392517e-14 | 1.754571e-12 | Bacteria | Proteobacteria | Gammaproteobacteria | Xanthomonadales | Xanthomonadaceae | Stenotrophomonas |
metro
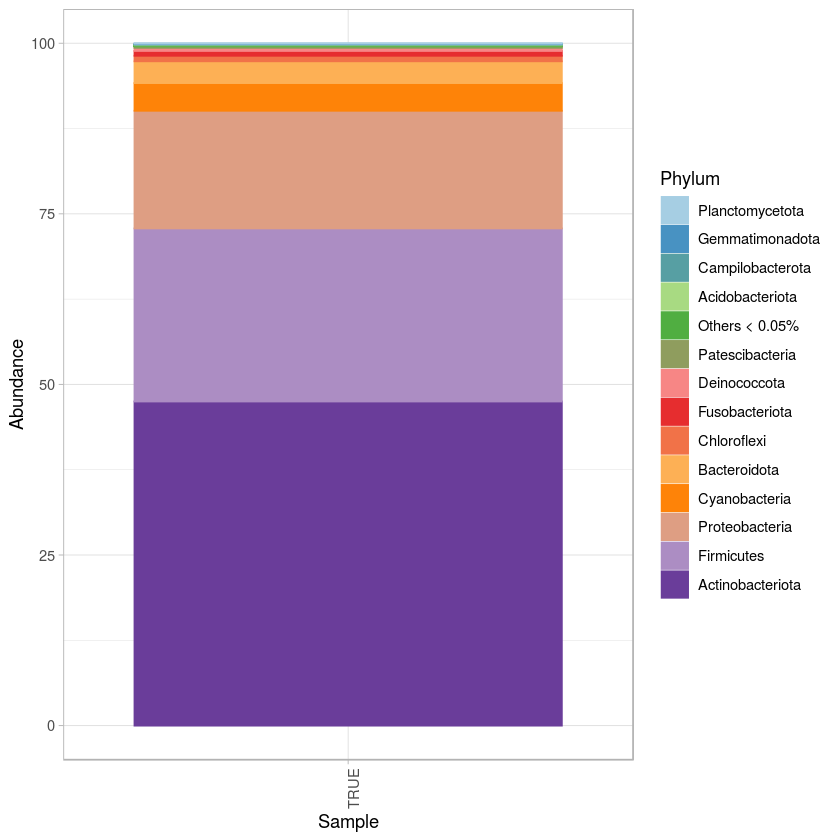
x4 <- tax_glom(metro, taxrank="Phylum")
metrotodos <- c("Pasamanos","Torniquete")
sample_data(x4)$todos = get_variable(x4, "sample_type") %in% metrotodos
mergeph1 <- merge_samples(x4, "todos")
mergeph1
x4 = transform_sample_counts(mergeph1,function(x) (x/sum(x))*100)
data <- psmelt(x4)
head(data)
data$Phylum <- as.character(data$Phylum)
data$Phylum[data$Abundance <= 0.05] <- "Others < 0.05%"
z <- factor(unique(data$Phylum))
z <- nlevels(z)
library(tidyverse)
pd <- data %>%
as_tibble %>%
mutate(Phylum = as.character(Phylum)) # %>%
# replace_na(list(Phylum = "unknown"))
Phylum_abun <- pd %>%
group_by(Phylum) %>%
summarize(Abundance = sum(Abundance)) %>%
arrange(Abundance)
Phylum_levels <- Phylum_abun$Phylum
pd0 <- pd %>%
mutate(Phylum = factor(Phylum, Phylum_levels))
p <- ggplot(pd0, aes(x = Sample, y = Abundance, fill= Phylum, color= Phylum)) + geom_bar(stat = "identity", position="stack") +
scale_fill_manual(values=(colorRampPalette(brewer.pal(10,"Paired"))(z))) +
scale_color_manual(values=(colorRampPalette(brewer.pal(10,"Paired"))(z))) + theme_light() +
theme(axis.text.x=element_text(angle=90, hjust=1)) #+ theme(legend.position="bottom") + guides(fill=guide_legend(nrow=2))
p
ggsave("phylum_allsamples_histogram.pdf")
phyloseq-class experiment-level object
otu_table() OTU Table: [ 22673 taxa and 47 samples ]
sample_data() Sample Data: [ 47 samples by 28 sample variables ]
tax_table() Taxonomy Table: [ 22673 taxa by 6 taxonomic ranks ]
phyloseq-class experiment-level object
otu_table() OTU Table: [ 40 taxa and 1 samples ]
sample_data() Sample Data: [ 1 samples by 29 sample variables ]
tax_table() Taxonomy Table: [ 40 taxa by 6 taxonomic ranks ]
| <th scope=col>OTU</th><th scope=col>Sample</th><th scope=col>Abundance</th><th scope=col>id</th><th scope=col>sample_location</th><th scope=col>sample_id</th><th scope=col>line_number</th><th scope=col>sample_type</th><th scope=col>date</th><th scope=col>time</th><th scope=col>⋯</th><th scope=col>latitude</th><th scope=col>longitude</th><th scope=col>people_affluence</th><th scope=col>Observed</th><th scope=col>Chao1</th><th scope=col>Shannon</th><th scope=col>Simpson</th><th scope=col>todos</th><th scope=col>Kingdom</th><th scope=col>Phylum</th> | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ASV_1 | TRUE | 47.5006996 | 24 | 17.68085 | 24 | 6.297872 | 1.510638 | 3.404255 | 19.85106 | ⋯ | NA | NA | 17859153 | 9903.553 | 15141.15 | 6.380126 | 0.9827077 | 1 | Bacteria | Actinobacteriota |
| ASV_2 | TRUE | 25.3627386 | 24 | 17.68085 | 24 | 6.297872 | 1.510638 | 3.404255 | 19.85106 | ⋯ | NA | NA | 17859153 | 9903.553 | 15141.15 | 6.380126 | 0.9827077 | 1 | Bacteria | Firmicutes |
| ASV_3 | TRUE | 17.2434928 | 24 | 17.68085 | 24 | 6.297872 | 1.510638 | 3.404255 | 19.85106 | ⋯ | NA | NA | 17859153 | 9903.553 | 15141.15 | 6.380126 | 0.9827077 | 1 | Bacteria | Proteobacteria |
| ASV_7 | TRUE | 4.0561480 | 24 | 17.68085 | 24 | 6.297872 | 1.510638 | 3.404255 | 19.85106 | ⋯ | NA | NA | 17859153 | 9903.553 | 15141.15 | 6.380126 | 0.9827077 | 1 | Bacteria | Cyanobacteria |
| ASV_90 | TRUE | 3.1905088 | 24 | 17.68085 | 24 | 6.297872 | 1.510638 | 3.404255 | 19.85106 | ⋯ | NA | NA | 17859153 | 9903.553 | 15141.15 | 6.380126 | 0.9827077 | 1 | Bacteria | Bacteroidota |
| ASV_182 | TRUE | 0.8116411 | 24 | 17.68085 | 24 | 6.297872 | 1.510638 | 3.404255 | 19.85106 | ⋯ | NA | NA | 17859153 | 9903.553 | 15141.15 | 6.380126 | 0.9827077 | 1 | Bacteria | Chloroflexi |
── Attaching packages ─────────────────────────────────────── tidyverse 1.2.1 ──
✔ tibble 2.1.3 ✔ purrr 0.2.5
✔ tidyr 0.8.2 ✔ dplyr 0.8.0.1
✔ readr 1.3.1 ✔ stringr 1.4.0
✔ tibble 2.1.3 ✔ forcats 0.4.0
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
Saving 6.67 x 6.67 in image
metro
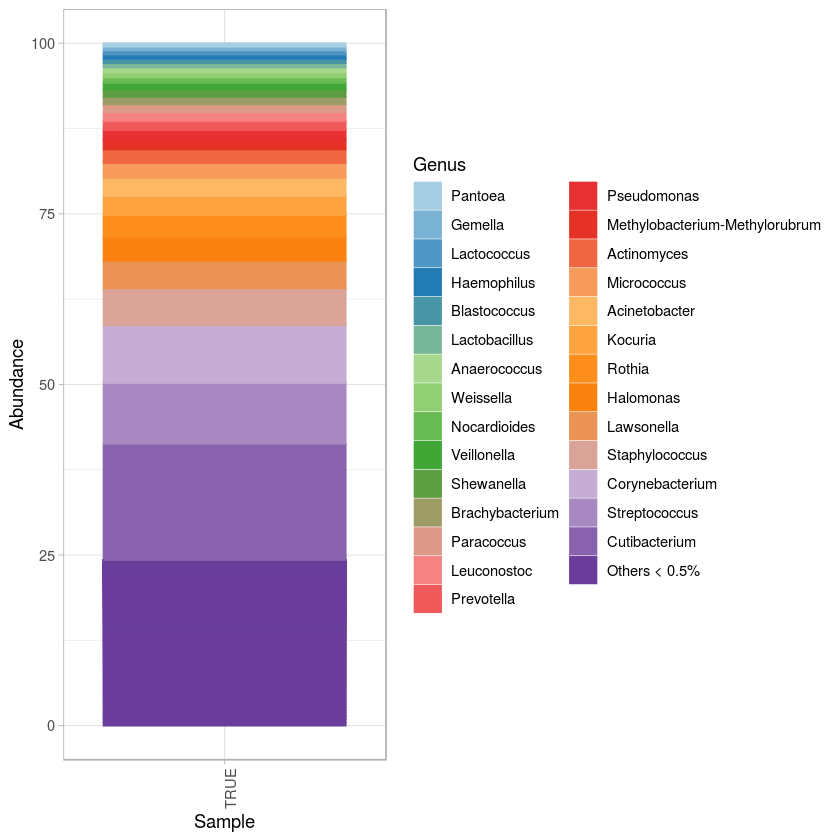
x4 <- tax_glom(metro, taxrank="Genus")
metrotodos <- c("Pasamanos","Torniquete")
sample_data(x4)$todos = get_variable(x4, "sample_type") %in% metrotodos
mergeph1 <- merge_samples(x4, "todos")
mergeph1
x4 = transform_sample_counts(mergeph1,function(x) (x/sum(x))*100)
data <- psmelt(x4)
head(data)
data$Genus <- as.character(data$Genus)
data$Genus[data$Abundance <= 0.5] <- "Others < 0.5%"
z <- factor(unique(data$Genus))
z <- nlevels(z)
z
library(tidyverse)
pd <- data %>%
as_tibble %>%
mutate(Genus = as.character(Genus)) # %>%
# replace_na(list(Phylum = "unknown"))
Genus_abun <- pd %>%
group_by(Genus) %>%
summarize(Abundance = sum(Abundance)) %>%
arrange(Abundance)
Genus_levels <- Genus_abun$Genus
pd0 <- pd %>%
mutate(Genus = factor(Genus, Genus_levels))
p <- ggplot(pd0, aes(x = Sample, y = Abundance, fill= Genus, color= Genus)) + geom_bar(stat = "identity", position="stack") +
scale_fill_manual(values=(colorRampPalette(brewer.pal(10,"Paired"))(z))) +
scale_color_manual(values=(colorRampPalette(brewer.pal(10,"Paired"))(z))) + theme_light() +
theme(axis.text.x=element_text(angle=90, hjust=1)) #+ theme(legend.position="bottom") + guides(fill=guide_legend(nrow=2))
p
ggsave("gemis_allsamples_histogram.pdf")
phyloseq-class experiment-level object
otu_table() OTU Table: [ 22673 taxa and 47 samples ]
sample_data() Sample Data: [ 47 samples by 28 sample variables ]
tax_table() Taxonomy Table: [ 22673 taxa by 6 taxonomic ranks ]
phyloseq-class experiment-level object
otu_table() OTU Table: [ 1252 taxa and 1 samples ]
sample_data() Sample Data: [ 1 samples by 29 sample variables ]
tax_table() Taxonomy Table: [ 1252 taxa by 6 taxonomic ranks ]
| <th scope=col>OTU</th><th scope=col>Sample</th><th scope=col>Abundance</th><th scope=col>id</th><th scope=col>sample_location</th><th scope=col>sample_id</th><th scope=col>line_number</th><th scope=col>sample_type</th><th scope=col>date</th><th scope=col>time</th><th scope=col>⋯</th><th scope=col>Chao1</th><th scope=col>Shannon</th><th scope=col>Simpson</th><th scope=col>todos</th><th scope=col>Kingdom</th><th scope=col>Phylum</th><th scope=col>Class</th><th scope=col>Order</th><th scope=col>Family</th><th scope=col>Genus</th> | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ASV_1 | TRUE | 17.029251 | 24 | 17.68085 | 24 | 6.297872 | 1.510638 | 3.404255 | 19.85106 | ⋯ | 15141.15 | 6.380126 | 0.9827077 | 1 | Bacteria | Actinobacteriota | Actinobacteria | Propionibacteriales | Propionibacteriaceae | Cutibacterium |
| ASV_2 | TRUE | 8.912395 | 24 | 17.68085 | 24 | 6.297872 | 1.510638 | 3.404255 | 19.85106 | ⋯ | 15141.15 | 6.380126 | 0.9827077 | 1 | Bacteria | Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | Streptococcus |
| ASV_6 | TRUE | 8.452160 | 24 | 17.68085 | 24 | 6.297872 | 1.510638 | 3.404255 | 19.85106 | ⋯ | 15141.15 | 6.380126 | 0.9827077 | 1 | Bacteria | Actinobacteriota | Actinobacteria | Corynebacteriales | Corynebacteriaceae | Corynebacterium |
| ASV_5 | TRUE | 5.407315 | 24 | 17.68085 | 24 | 6.297872 | 1.510638 | 3.404255 | 19.85106 | ⋯ | 15141.15 | 6.380126 | 0.9827077 | 1 | Bacteria | Firmicutes | Bacilli | Staphylococcales | Staphylococcaceae | Staphylococcus |
| ASV_4 | TRUE | 4.045221 | 24 | 17.68085 | 24 | 6.297872 | 1.510638 | 3.404255 | 19.85106 | ⋯ | 15141.15 | 6.380126 | 0.9827077 | 1 | Bacteria | Actinobacteriota | Actinobacteria | Corynebacteriales | Corynebacteriaceae | Lawsonella |
| ASV_3 | TRUE | 3.516355 | 24 | 17.68085 | 24 | 6.297872 | 1.510638 | 3.404255 | 19.85106 | ⋯ | 15141.15 | 6.380126 | 0.9827077 | 1 | Bacteria | Proteobacteria | Gammaproteobacteria | Oceanospirillales | Halomonadaceae | Halomonas |
29
Saving 6.67 x 6.67 in image
x4
phyloseq-class experiment-level object
otu_table() OTU Table: [ 1252 taxa and 1 samples ]
sample_data() Sample Data: [ 1 samples by 29 sample variables ]
tax_table() Taxonomy Table: [ 1252 taxa by 6 taxonomic ranks ]